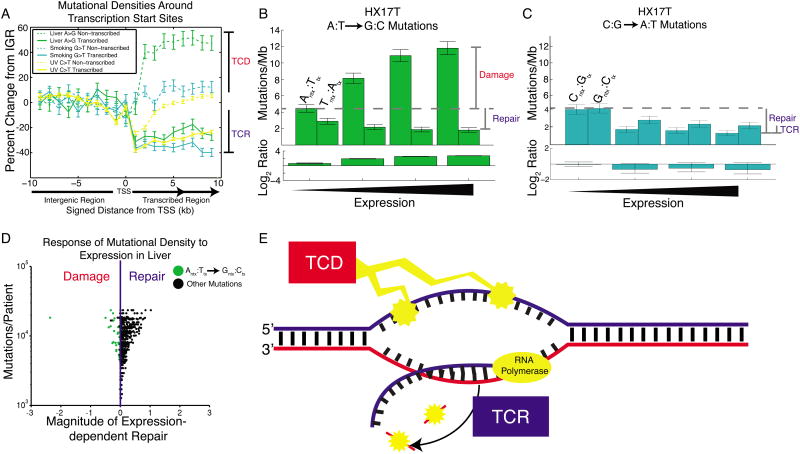
Figure 7. Transcription-coupled damage in liver cancer.
(A) Mutational densities in the vicinity of promoters. When crossing from non-transcribed regions (IGR) to transcribed regions, mutational densities on the transcribed strand fall, reflecting transcription-coupled repair (TCR). On the non-transcribed strand there is usually little change from IGR levels, with the notable exception of liver cancer, where mutational densities increase from IGR levels, consistent with transcription-coupled damage (TCD). (B) Liver cancer patient HX17T shows a dramatic expression-dependent increase in A→G mutational densities on the non-transcribed strand only. (C) In the same patient, G→T mutational densities show only the usual expression-dependent decrease, on both strands. (D) Most liver patients show dominant TCR. However, for A→G mutations on the non-transcribed strand (green dots), some show the opposite trend, reflecting dominant TCD. The leftmost dot is patient HX17T. (E) TCD damages the non-transcribed strand, exposed as ssDNA during transcription. TCR repairs the transcribed strand. Both of these processes contribute to T-class asymmetry.