Abstract
Nasal olfactory mucosa mesenchymal stem cells (OM-MSCs) have the ability to promote regeneration in the nervous system in vivo. Moreover, with view to the potential for clinical application, OM-MSCs have the advantage of being easily accessible from patients and transplantable in an autologous manner, thus eliminating immune rejection and contentious ethical issues. So far, most studies have been focused on the role of OM-MSCs in central nervous system replacement. However, the secreted proteomics of OM-MSCs have not been reported yet. Here, proteins secreted by OM-MSCs cultured in serum-free conditions were separated on SDS-PAGE and identified by LC-MS/MS. As a result, a total of 274 secreted proteins were identified. These molecules are known to be important in neurotrophy, angiogenesis, cell growth, differentiation, and apoptosis, and inflammation which were highly correlated with the repair of central nervous system. The proteomic profiling of the OM-MSCs secretome might provide new insights into their nature in the neural recovery. However, proteomic analysis for clinical biomarkers of OM-MSCs needs to be further studied.
1. Introduction
Repairing the central nervous system (CNS) has always been a challenge of science prompting innovative strategies. There is an old view that the nerves have little potential to rebirth according to cognitive demands of the environment [1]. Even though neural stem cells (NSCs) were capable of self-renewal and generating the main phenotypes (neurons, astrocytes, and oligodendrocytes) of CNS cells in vitro and in vivo [2], endogenous NSCs usually fail to remedy the deleterious consequences of severe trauma or neurodegenerative diseases [3–5]. Therefore, exogenous cell therapy has been proposed as a promising approach to treat a variety of neurological diseases [6].
Among the candidate cells for clinical cell therapy, mesenchymal stem cells (MSCs) are considered to be an ideal cell type for nerve repair and clinical applications [7, 8]. MSCs could be obtained from a variety of tissues, including bone marrow, umbilical cord, umbilical cord blood, adipose tissue, synovium, skeletal muscle, and deciduous teeth [9]. Among them, bone marrow mesenchymal stem cells are well-studied. However, it is still faced with many restrictions, including high susceptibility to viral exposure, the invasiveness of marrow collection, the rarity of MSCs in the marrow, cell proliferation ability, and cell differentiation capacity decrease with aging [10–12]. Thus, researchers have never stopped looking for new seed cells for clinical cell transplantation.
Recently, nasal olfactory mucosa mesenchymal stem cells (OM-MSCs) stand as a promising competitor for therapeutic application [13–15]. It has the advantages of easy accession, advantageous localization, and high versatility [16]. Most importantly, OM-MSCs could be used for autologous transplantation. Presently, these cells have been successfully used in different mammals models, including myocardial infarct [17], spinal cord trauma [18–20], hippocampal lesions [21], Parkinson's disease [22], and cochlear damage [23, 24]. Although OM-MSCs present the advantage of potential usefulness for autologous transplantation, the detailed mechanisms are only partially understood.
It has been well-accepted that the reparative effect of OM-MSCs is due to not only its differentiation capacity, but also the paracrine factors [25, 26]. These paracrine factors might play important roles in creating a supportive microenvironment for cell survival and differentiation, the activation of endogenous NSCs, reduction of inflammatory reaction, and inducement of angiogenesis [27]. Therefore the secretome of OM-MSCs could be very useful to uncover the active ingredients for further study on its mechanism and clinical application.
Here, the secretome of OM-MSCs was investigated by SDS-PAGE and LC-MS. A total of 274 secreted proteins were identified. These molecules are known to be important in neurotrophy, angiogenesis, cell growth, differentiation, and apoptosis, and inflammation indicating that OM-MSC transplantation could significantly improve the repair of CNS. The proteins presented in this study represent the first secretome profiling of OM-MSC, which may provide insight into its functions in nerve repairment.
2. Materials and Methods
2.1. Ethics Statement
Human nasal mucosa biopsies were collected from 4 individuals, 2 males and 2 females aged 24 to 49 years, at the Second Affiliated Hospital of Hunan Normal University (Changsha, China); investigations were approved by the ethical committee of Hunan Normal University, China. Informed consent was given by each individual participating in the study, in accordance with the Helsinki convention (1964). Inferior turbinate tissues were discarded from 4 patients undergoing septoplasty/polypectomy surgery. The patients with history of facial trauma were excluded.
2.2. Tissue Culture
Following published protocol for the cultivation of human nasal lamina propria MSCs [16], samples were collected at the root of the medial aspect of the middle turbinate, washed 3 times at room temperature with antibiotic-antimycotic solution (Invitrogen, Carlsbad, CA), and then cut into 1 mm3 to 2 mm3 pieces with a thickness ranging from 200 to 500 μm. The pieces were placed into a culture dish that was covered with a sterilized glass cover slide. Dulbecco's modified Eagle's medium (DMEM; Invitrogen) containing 10% fetal bovine serum (FBS) was added, and the tissues were incubated at 37°C in 5% CO2. The culture medium was changed every 2 or 4 days. Seven to eight days after, stem cells will begin to invade the culture dish and after 2-3 weeks they should be confluent. When confluency is reached, passage and transfer the cells to culture flasks. When almost confluent, cells were again harvested and were then plated in sphere-forming conditions. Cell density appeared to be important for robust sphere formation. Cells were plated at 4 × 105 to 6 × 105 cells per poly-L-lysine coated T25 flask in DMEM/F12 supplemented with 50 ng/mL fibroblast growth factor 2 (R&D Systems, Minneapolis, MN) and 50 ng/mL epidermal growth factor (R&D Systems). Medium was added every 2 to 4 days. Loosely adherent spheres were harvested by trituration and brief Accutase (Invitrogen) treatment for immunochemical analysis.
2.3. Immunofluorescent Staining
Immunofluorescence was carried out to analyze nestin, STRO-1, and microtubule-associated protein-2 (MAP-2) expression using standard protocols. After fixation and washing, cultures were blocked with 10% normal goat serum in 0.3% Triton X-100 for 1 hr at room temperature and incubated with the primary antibody at 4°C overnight. The following primary antibodies were used: monoclonal rabbit anti-nestin (1 : 250, Abcam, Cambridge, UK) and mouse anti-STRO-1 (1 : 250, Sigma-Aldrich, CA, USA) for OM-MSCs and monoclonal anti-MAP-2 (1 : 250, Abcam) for neuron-like cells. Then cultures were incubated with the appropriate conjugated secondary antibody for 1 hr, at room temperature in dark conditions. The following secondary antibodies were used: goat anti-mouse IgG conjugated with AlexaFluor 488 (1 : 500, Santa Cruz, CA, USA) and donkey anti-rabbit IgG conjugated with AlexaFluor 647 (1 : 500, Santa Cruz). After several washes in PBS, cultures were counterstained with DAPI (4′,6-diamidino-2-phenyl-indole; Roche, Germany) for 5 min and mounted with an antifading medium (Beyotime, Hangzhou, China). Images were taken with a fluorescence microscopy (Leica DM 2000, Germany).
2.4. Flow Cytometry Analysis
For surface protein expression, OM-MSCs were plated into a test tube (Becton Dickinson, NJ, USA) at a density of 1 × 105/mL and washed three times with wash buffer (0.1% FBS/PBS). The cells were incubated for 40 min with saturating concentrations of fluorescent-conjugated monoclonal antibodies CD34, CD45, CD73, CD90, and CD105. Anti-CD34 was conjugated to ECD, anti-CD45 was conjugated to PC7, anti-CD73 and anti-CD90 were conjugated to FITC, and anti-CD105 was conjugated to PE. After washing, cell fluorescence signals were determined immediately using flow cytometry with a FACS Caliber instrument (Becton Dickinson, CA, USA). The analysis was performed using Cell Quest Software (Becton Dickinson, CA, USA) [28, 29].
2.5. Osteogenic and Adipogenic Differentiation
The ability of OM-MSCs to differentiate into adipogenic and osteogenic lineages was assayed on cells plated at 102 to 103 cells/cm2 in DMEM/F12, 10% FBS, and penicillin/streptomycin in 6-well plates. 24 hours later, using StemPro osteogenesis and adipogenesis differentiation kits (Invitrogen), induction factors were added to 3 wells and 3 wells kept as controls. Cells were fed every other day for 14 days and bone and fat were detected using alizarin red S stain, alkaline phosphatase stain, and Oil red O stain.
2.6. Neurogenic Differentiation
To induce neurogenesis, single OM-MSCs were placed in nonadherent 6-well culture plates with a density of 100,000 cells per mL for 24–48 hours in the DMEM/F12 medium supplemented with 10% FBS, penicillin/streptomycin. Then, the medium was replaced with DMEM/F12 supplemented with 500 nM retinoic acid (Sigma-Aldrich) for 12 days. The differentiated cells were used for western blot and immunocytochemistry of immature to mature neural markers such as Neuronal Class III β-tubulin-1 (β-TUB), MAP-2, nestin, and glial fibrillary acidic protein (GFAP).
2.7. Protein Extraction
Proteins of OM-MSCs were harvested by washing cultured and acutely dissociated OM-MSCs in cold PBS and lysing with 500 μL of 1x SDS-PAGE sample buffer. Samples were incubated on ice and stored at −20°C.
To obtain secreted protein, approximately 80% confluent OM-MSC cultures were rinsed three times with PBS and incubated overnight in DMEM/F12 conditioned medium. The conditioned medium was collected and centrifuged at 500 g. The supernatant was filtered using a 0.2 μm filter and placed in dialysis cassettes with molecular weight cut-off of 3500 (Pierce), dialyzed against three changes of 10 volumes of 1 mM ammonium bicarbonate solution. The dialyzed CM was lyophilized overnight and then resuspended in 1x SDS sample buffer.
Total proteins were quantified using the Bradford method.
2.8. SDS-PAGE and Western Blotting
SDS-PAGE protein gels were performed according to standard procedures. Equal amount (100 μg) of proteins was boiled for 5 min and centrifuged at 12,000 g for 10 min at 4°C. Samples were subjected to SDS-PAGE using 11.5% separation gel and 4.8% stacking gel. After electrophoresis, the gel was stained using coomassie brilliant blue. To prepare samples for western blotting, 20–50 μg samples were separated on SDS-PAGE, transferred to PVDF membrane (Millipore, MA, USA), and then probed with various antibodies overnight at 4°C. The dilution rates of primary antibodies were as follows: anti-β-TUB (1 : 200), anti-GFAP (1 : 400), anti-MAP-2 (1 : 400), and anti-nestin (1 : 300). Next, the membrane was incubated with HRP conjugated anti-rabbit or anti-mouse IgG, and after fully washing with TBS the blot was visualized using ChemiDoc XRS imaging system (Bio-Rad, CA, USA).
2.9. Tryptic Digestion and MS/MS Analysis
The stained gel was sliced into fifteen bands of equal size, and a standard protocol was performed for in-gel digestion as described before [30, 31]. The trypsinized sample was lyophilized, dissolved in 0.1% formic acid, and analyzed using an LTQ mass spectrometer (Thermo Finnigan, San Jose, CA) coupled with an Agilent 1200 capillary system (Agilent, Waldbronn, Germany). Mobile phase A (0.1% formic acid in H2O) and mobile phase B (0.1% formic acid in acetonitrile) were selected. A linear 65 min gradient was achieved from 3% to 60% B at a flow rate of 0.3 μL/min. The eluted peptide ions were then detected by the LTQ mass spectrometer equipped with the nano-ESI source. The electrospray voltage was 1.9 kV, and 35% normalized collision energy was used for MS/MS. Data-dependent MS/MS spectra were obtained in which the five most abundant spectra from the full MS scan were selected for fragmentation. The following are the dynamic exclusion settings: the repeat count was set to 1, the repeat duration was 30 s, the dynamic exclusion duration was set to 180 s, the exclusion mass width was 1.5 Da, and the list of dynamic exclusion was 50.
2.10. Database Searching and Bioinformatics Analysis
Raw MS data were processed by the TurboSEQUEST program in the Bioworks Browser software suite (version 3.1, Thermo Electron, San Jose, CA). The following SEQUEST search parameters were used: precursor mass tolerance was set at 15 ppm and 0.8 Da for MS/MS fragments. The matched peptides were allowed up to 1 missed cleavage and oxidation on Met (+16 Da) as variable modification while carbamidomethylation on Cys (+57 Da) as fixed modification. Peptides whose ions scores exceeded the threshold (p < 0.05, 95% confidence level) were selected and a list of proteins that had a ProteinProphet probability greater than 0.99 and also had more than one unique peptide were obtained.
The accession number of each identified protein was loaded to the gene ontology (GO) classification system (http://www.geneontology.org/). The proteins were classified into the different cellular component, molecular function, and biological process. General functions analysis was performed with the tools on the Swiss-Prot database (http://www.uniprot.org/), respectively.
3. Results
3.1. Isolation of Human Olfactory Mucosa (OM) Biopsies
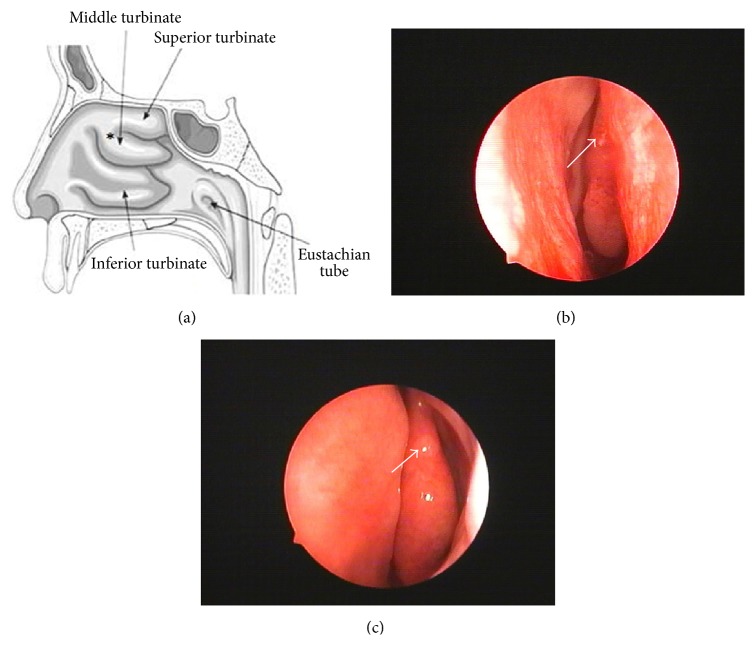
Before collection, certain diseases, especially nasal polyps and/or active sinus infection, are necessary to be excluded. The protocol was designed according to IRB after passing the ethical committee and all volunteers signed the informed consent for the use of intranasal biopsy samples for research. Then tissue samples were obtained from the root of the medial aspect of the middle turbinate undergoing endoscopic nasal surgery (Figure 1(a)). Usually OM of patients could heal within a month after injury. Moreover, there is no effect on the patients' sense of smell through olfactory measurement (Figures 1(b) and 1(c)).
Figure 1.
Otolaryngologic image of the nasal cavity. Schematic of the right nasal cavity, ∗: at the root of the middle turbinate (a), endoscopic visualization of the obtaining (b), and endoscopic pictures after one month (c).
3.2. Characterization and Sphere-Forming Capabilities of OM-MSCs
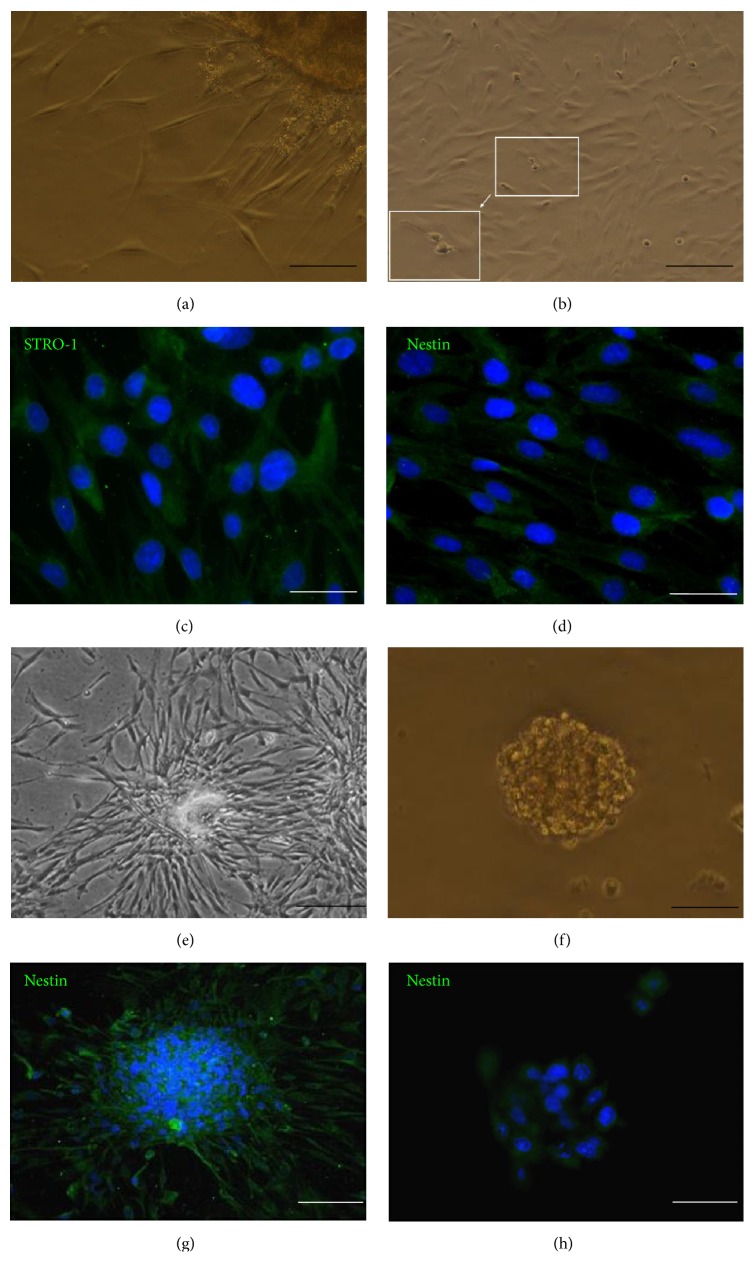
To validate that the correct population of cells had been harvested from OM, we evaluated whether it was possible to isolate stem cells. Following the protocols for the generation of OM-MSC cultures, we successfully derived cells exhibiting a mesenchymal stem cell like morphology (Figures 2(a) and 2(b)). After 6 to 8 days in culture, adherent cells migrated from the explants and most cells became spindle shaped (Figure 2(a)). After passaging, the cells grow rapidly and the nuclear disintegration and cell division could be observed (Figure 2(b)), showing active proliferation capacity. Moreover, purified cells were immunopositive for STRO-1 and nestin, which are the characteristic markers of human OM-MSCs (Figures 2(c) and 2(d)) [14]. After culturing for 14 days, as expected, cells tended to aggregate to loosely attached or floating spheres (Figures 2(c) and 2(d)) in which the signal of nestin is strong (Figures 2(e) and 2(f)). Indeed, even though derived from very small pieces of tissue, millions of cells could be obtained within a month easily.
Figure 2.
Characterization and sphere-forming capabilities of OM-MSCs. Adherent cells migrated from the explants and most cells became spindle shaped (a), the rapidly grown cells within nuclear disintegration and cell cleavage (b), immunocytochemistry of the characteristic markers of human OM-MSCs: STRO-1 ((c) and (g)) and nestin ((d) and (h)). After cultivating in the appropriate culture conditions, as expected, cells tended to aggregate; many loosely attached or floating spheres appeared ((e) and (f)). Neurospheres can be expressed as nestin throughout the whole process; nuclei are counterstained with DAPI (blue). Scale bars: (a) 100 μm; (b) 200 μm, white box: 100 μm; (c) and (d) 50 μm; (e) and (g) 200 μm; (f) and (h) 50 μm. DAPI = 4′,6-diamidino-2-phenylindole; MSC = mesenchymal stem cell.
3.3. Surface Marker Expression
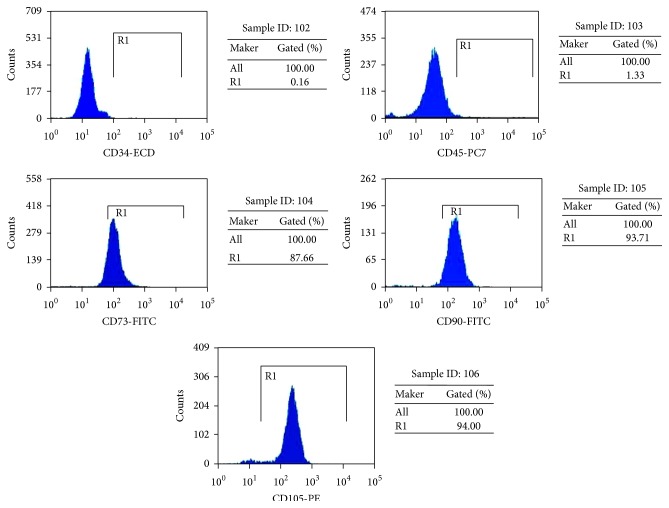
To determine the immunophenotyping profile of OM-MSCs, we used flow cytometry to assess different cell surface antigens. Flow cytometric analysis of these cells showed that they express the MSC markers CD73, CD90, and CD105, but not hematopoietic cells markers CD34 and CD45 (Figure 3). These results suggested that these cells are mainly composed of MSCs.
Figure 3.
Surface marker expression. Flow cytometric analysis of these cells showed that they express the MSC markers CD73, CD90, and CD105, but not CD34 and CD45, which are characteristic of hematopoietic cells.
3.4. OM-MSCs Differentiate into Osteoblasts, Adipocytes, and Neurons In Vitro
Researches have shown that MSCs could differentiate into bone, fat, cartilage, and other types of cells; thus the pluripotency of the OM-MSCs was measured.
3.4.1. In Vitro Adipogenic Assay
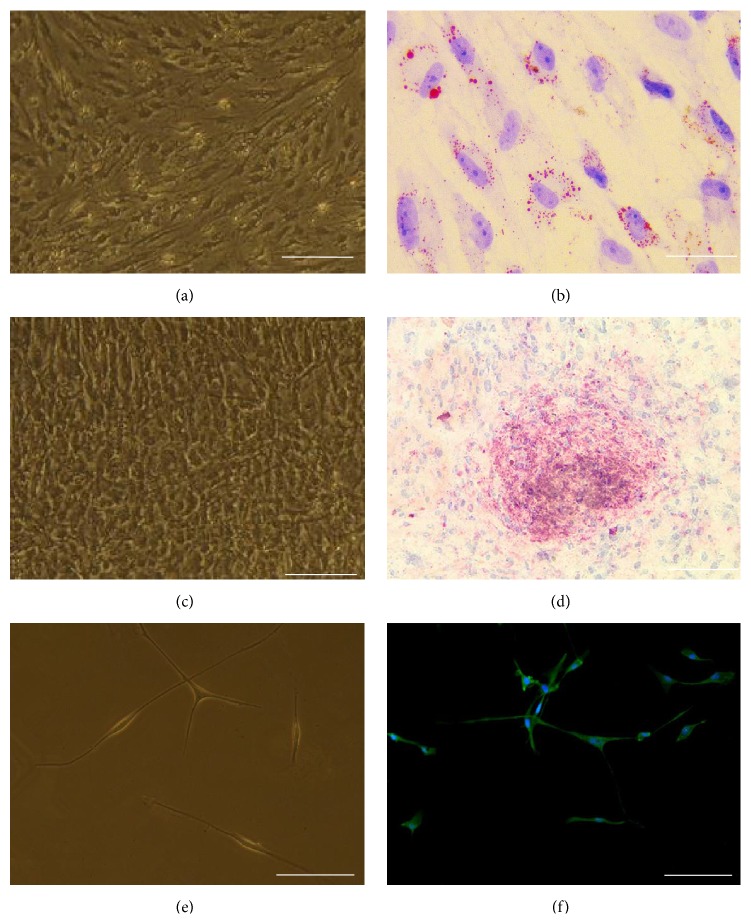
To test if OM-MSCs have the capacity to differentiate into fat cells, osteogenesis differentiation factors were added. After induction for 18 days, the OM-MSCs became shorter and bigger. Highly refractive vacuoles were observed under phase contrast microscope (Figure 4(a)). After Oil red O staining, a large number of lipid droplets in the cytoplasm were stained red (Figures 4(a) and 4(b)).
Figure 4.
OM-MSCs differentiate in vitro into osteoblasts, adipocytes, and neurons. Adipogenic, osteogenic, and neuron differentiation was observed and assessed by Oil red O (ORO), alizarin red staining (ARS), or MAP-2 in induction media. (a) Adipocyte cellular morphology in 2 weeks; (b) lipid droplets that showed red color after ORO staining; (c) osteocyte cellular morphology in 3 weeks; (d) ARS that verified the formation of large calcium deposits. (e) Neuron cellular morphology in 12 days; (f) neuron-like cells that can be expressed MAP-2. Nuclei are counterstained with DAPI (blue). Scale bars: (a) and (c) 200 μm; (b) and (d) 100 μm; (e) and (f) 50 μm. DAPI = 4′,6-diamidino-2-phenylindole; MSC = mesenchymal stem cell.
3.4.2. In Vitro Osteogenic Assay
To test if OM-MSCs have the capacity to differentiate into bone cells, adipogenesis differentiation factors were added, too. After induction, OM-MSCs became cubical and showed tendency to aggregate. The alizarin red S was positively stained after osteogenic induction for 3 weeks, suggesting that the OM-MSCs have the potential to differentiate into osteoblasts in vitro (Figures 4(c) and 4(d)).
3.4.3. In Vitro Neurogenic Assay
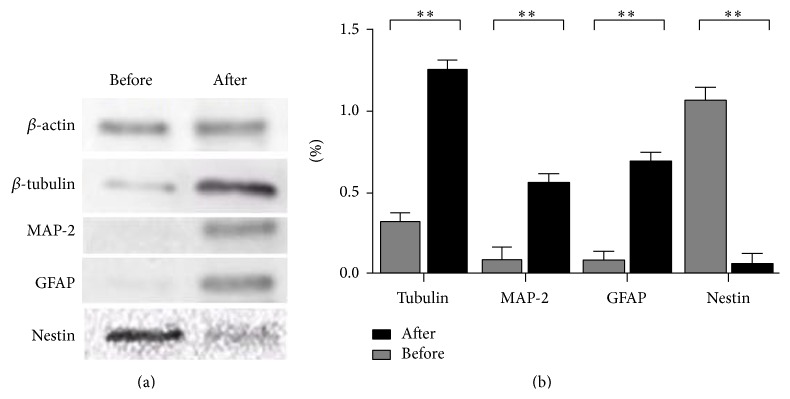
To test if OM-MSCs have the capacity to differentiate into neuronal-like cells, cells were harvested and reseeded into differentiation conditions. After induction with retinoic acid for 12 days, the typical morphology of the neurites was presented and the neuron-like cell marker MAP-2 expression was revealed by immunofluorescence (Figures 4(e) and 4(f)). The following western blotting showed that OM-MSCs were expressing neuron-like cell marker MAP-2, β-TUB, and the glia marker GFAP. At the same time, the expression level of stem cell marker nestin was observably decreased (Figure 5).
Figure 5.
Western blot assay of OM-MSCs neuronal induction for 1 and 2 weeks. The differentiation of neuron-like cells from mesenchymal stem cells (MSCs) was verified by western blot. After induced, the neuron marker β-tubulin and MAP-2 were highly increased, and the stem cell marker, nestin, indicated significant downregulation.
3.5. Separation and Identification of Secreted Proteins
The secreted proteins of OM-MSCs were prefractionated via 1D-SDS-PAGE (Figure 6(a)). The gels were cut into 15 parts equally and trypsinized by in-gel digestion. Trypsinized peptides were analyzed by MS in triplicate and the results were processed using a sequential analysis as described in the proteomic data processing section. Shotgun proteomics with these rigorous conditions identified a total of 274 nonredundant proteins in OM-MSC conditioned medium. The entire list of identified proteins was provided in Table S1 (http://1000eb.com/1g25l) (see Supplementary Material available online at http://dx.doi.org/10.1155/2016/1243659).
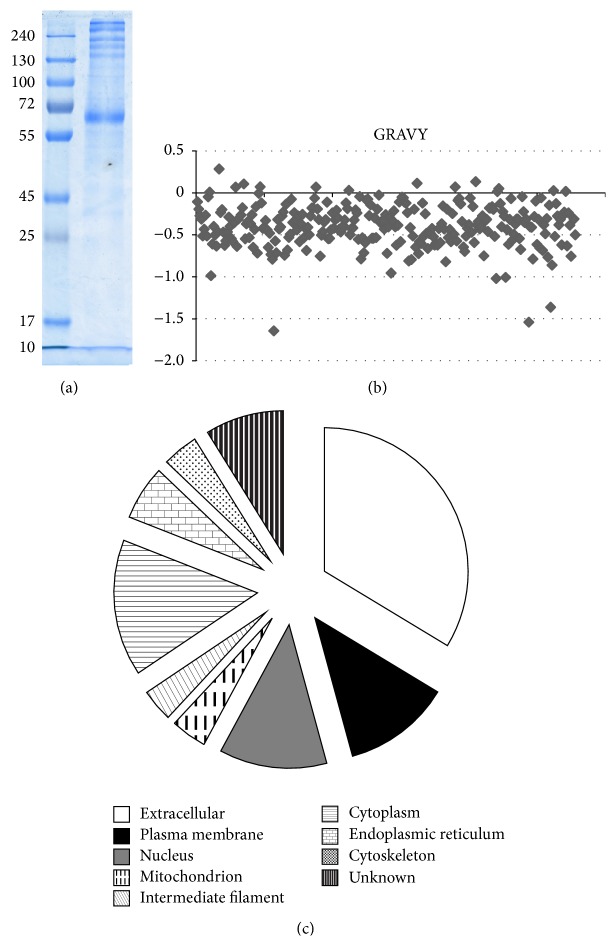
Figure 6.
The SDS-PAGE analysis of the proteins isolated from OM-MSC conditioned medium and the hydrophobicity and primary subcellular localization of the identified proteins in OM-MSC conditioned medium. SDS-PAGE analysis of the secreted proteins of OM-MSC (a). The GRAVY values varied from −1.647 to 0.282 and 261 (94.5%) of them are negative, showing these proteins were hydrophilic and might be unidentified secreted proteins and functioned as growth factors or signaling molecules (b). Of the 276 identified proteins from OM-MSC conditioned medium, 175 (63.4%) were recognized as extracellular proteins through GO cellular component assignment. Classification by cellular localization is redundant since a protein can be classified in more than one compartment (c).
The hydrophobicity property of proteins was expressed as the GRAVY index and the ProtParam tool at ExPASy was used to calculate the GRAVY values of proteins. The proteins exhibiting positive GRAVY values were recognized to be hydrophobic and those with negative values were deemed hydrophilic. As for the identified extracellular proteins in this study, the GRAVY values varied from −1.647 to 0.282 and 261 (94.5%) of them are negative (Figure 6(b)), showing that these proteins were hydrophilic.
Of the 276 identified proteins from OM-MSC conditioned medium, 175 (63.4%) were recognized as extracellular proteins through GO cellular component assignment (Figure 6(c)). Other identified proteins included plasma membrane (63, 22.8%), nucleus (63, 22.8%), mitochondrion (21, 7.6%), intermediate filament (19, 6.9%), cytoplasm (80, 29.0%), endoplasmic reticulum (32, 11.6%), cytoskeleton (21, 7.6%), and unknown (46, 16.7%). Classification by cellular localization is redundant since a protein can be classified into more than one compartment.
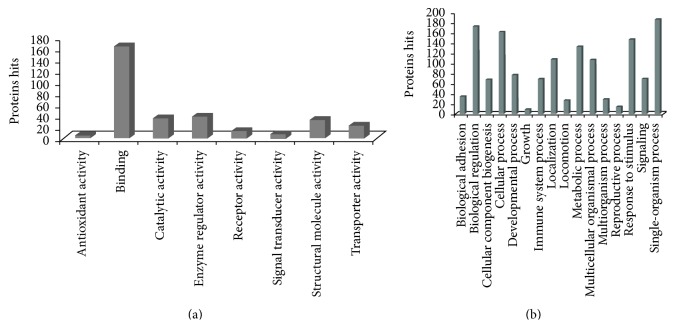
The molecular functions of the identified secreted proteins were composed of several categories, such as antioxidant activity, binding, catalytic activity, enzyme regulator activity, receptor activity, signal transducer activity, structural molecule activity, and transporter activity.
3.6. Biological Processes and Function Annotation
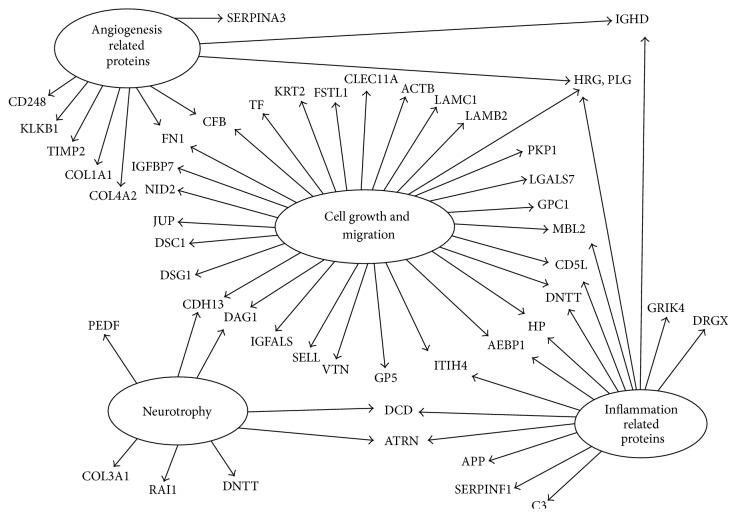
To investigate whether the secreted factors could function in the tissues repairment, GO analysis was conducted. Significantly higher frequencies of genes (p < 0.05) were associated with 16 biological processes (Figure 7). These proteins were most frequently involved in metabolism, defense response, signaling, and tissue differentiation. Proteins that have the function annotations in Swiss-Prot database were classified into 4 groups named related to neurotrophy, related to blood circulation, related to cell growth, differentiation, and apoptosis, and proteins associated with inflammation (Table 1, Figure 8).
Figure 7.
Molecular functions and biological processes of extracellular proteins identified in OM-MSC conditioned medium. The molecular functions of the identified secreted proteins were composed of several categories, such as antioxidant activity, binding, catalytic activity, enzyme regulator activity, receptor activity, signal transducer activity, structural molecule activity, and transporter activity (a). It indicated that the main functions of secreted proteins are maintaining the homeostasis of extracellular matrix, transmitting signals, and so on. Significantly higher frequencies of genes (p < 0.05) were associated with 16 biological processes (b). These proteins were most frequently involved in metabolism, defense response, signaling, and tissue differentiation.
Table 1.
Function classification of OM-MSCs secreted proteins.
| Cell growth/proliferation/differentiation/apoptosis/survival/adhesion/migration | ||
|---|---|---|
| Accession | Gene name | Protein description |
| P02787 | TF | Serotransferrin |
| Q14118 | DAG1 | Dystroglycan |
| Q8IUX7 | AEBP1 | Adipocyte enhancer-binding protein 1 |
| P00751 | CFB | Complement factor B |
| P35908 | KRT2 | Keratin, type II cytoskeletal 2 epidermal |
| Q12841 | FSTL1 | Follistatin-related protein 1 |
| Q9Y240 | CLEC11A | C-type lectin domain family 11 member A |
| P60709 | ACTB | Actin, cytoplasmic 1 |
| P11047 | LAMC1 | Laminin subunit gamma-1 |
| P55268 | LAMB2 | Laminin subunit beta-2 |
| P00738 | HP | Haptoglobin |
| P04053 | DNTT | DNA nucleotidylexotransferase |
| P14923 | JUP | Junction plakoglobin |
| Q13835 | PKP1 | Plakophilin-1 |
| Q14624 | ITIH4 | Inter-alpha-trypsin inhibitor heavy chain H4 |
| O43866 | CD5L | CD5 antigen-like |
| P11226 | MBL2 | Mannose-binding protein C |
| P04196 | HRG | Histidine-rich glycoprotein |
| P17936 | IGFBP3 | Insulin-like growth factor-binding protein 3 |
| P18065 | IGFBP2 | Insulin-like growth factor-binding protein 2 |
| P22692 | IGFBP4 | Insulin-like growth factor-binding protein 4 |
| P35052 | GPC1 | Glypican-1 |
| P47929 | LGALS7 | Galectin-7 (Gal-7) |
| Q6E0U4 | DMKN | Dermokine |
| Q7Z5J4 | RAI1 | Retinoic acid-induced protein 1 |
| P00747 | PLG | Plasminogen |
| Q71UI9 | H2AFV | Histone H2A.V |
| Q66K74 | MAP1S | Microtubule-associated protein 1S |
| P10909 | CLU | Clusterin |
| Q9UBP9 | GULP1 | PTB domain-containing engulfment adapter protein 1 |
| P55290 | CDH13 | Cadherin-13 |
| P02751 | FN1 | Fibronectin |
| Q12913 | PTPRJ | Receptor-type tyrosine-protein phosphatase eta |
| P40197 | GP5 | Platelet glycoprotein V |
| P04004 | VTN | Vitronectin |
| P14151 | SELL | L-selectin |
| P35858 | IGFALS | Insulin-like growth factor-binding protein complex acid labile subunit |
| Q02413 | DSG1 | Desmoglein-1 |
| Q08554 | DSC1 | Desmocollin-1 |
| Q14112 | NID2 | Nidogen-2 |
| Q16270 | IGFBP7 | Insulin-like growth factor-binding protein 7 |
| Q86SJ6 | DSG4 | Desmoglein-4 |
| Q9UI47 | CTNNA3 | Catenin alpha-3 |
| Q9Y6C2 | EMILIN1 | EMILIN-1 |
| F5GY03 | SPARC | SPARC |
|
| ||
| Angiogenesis and blood circulation | ||
| P00751 | CFB | Complement factor B |
| P04196 | HRG | Histidine-rich glycoprotein |
| P00747 | PLG | Plasminogen |
| P00734 | F2 | Prothrombin |
| P05154 | SERPINA5 | Plasma serine protease inhibitor |
| P02763 | ORM1 | Alpha-1-acid glycoprotein 1 |
| P02774 | GC | Vitamin D-binding protein |
| P00748 | F12 | Coagulation factor XII |
| P01008 | SERPINC1 | Antithrombin-III |
| P01009 | SERPINA1 | Alpha-1-antitrypsin |
| P01019 | AGT | Angiotensinogen |
| P01042 | KNG1 | Kininogen-1 |
| P02749 | APOH | Beta-2-glycoprotein 1 |
| P02768 | ALB | Serum albumin |
| P05155 | SERPING1 | Plasma protease C1 inhibitor |
| P07359 | GP1BA | Platelet glycoprotein Ib alpha chain |
| P12259 | F5 | Coagulation factor V |
| P68871 | HBB | Hemoglobin subunit beta |
| Q9UNN8 | PROCR | Endothelial protein C receptor |
| P02649 | APOE | Apolipoprotein E |
| P08572 | COL4A2 | Collagen alpha-2(IV) chain |
| P01011 | SERPINA3 | Alpha-1-antichymotrypsin |
| P02452 | COL1A1 | Collagen alpha-1(I) chain |
| P03952 | KLKB1 | Plasma kallikrein |
| P16035 | TIMP2 | Metalloproteinase inhibitor 2 |
| Q9HCU0 | CD248 | Endosialin |
| P02751 | FN1 | Fibronectin |
|
| ||
| Inflammation/immune regulation | ||
| Q8IUX7 | AEBP1 | Adipocyte enhancer-binding protein 1 |
| P00738 | HP | Haptoglobin |
| P04053 | DNTT | DNA nucleotidylexotransferase |
| P81605 | DCD | Dermcidin |
| P01024 | C3 | Complement C3 |
| P01880 | IGHD | Ig delta chain C region |
| O43866 | CD5L | CD5 antigen-like |
| P11226 | MBL2 | Mannose-binding protein C |
| P04196 | HRG | Histidine-rich glycoprotein |
| P01871 | IGHM | Ig mu chain C region |
| P01876 | IGHA1 | Ig alpha-1 chain C region |
| P01877 | IGHA2 | Ig alpha-2 chain C region |
| P05546 | SERPIND1 | Heparin cofactor 2 |
| P07357 | C8A | Complement component C8 alpha chain |
| P0C0L4 | C4A | Complement C4-A |
| P10643 | C7 | Complement component C7 |
| P13671 | C6 | Complement component C6 |
| P15814 | IGLL1 | Immunoglobulin lambda-like polypeptide 1 |
| P19652 | ORM2 | Alpha-1-acid glycoprotein 2 |
| P48740 | MASP1 | Mannan-binding lectin serine protease 1 |
| Q6UXS9 | CASP12 | Inactive caspase-12 |
| P00747 | PLG | Plasminogen |
| P00734 | F2 | Prothrombin |
| P05154 | SERPINA5 | Plasma serine protease inhibitor |
| P02763 | ORM1 | Alpha-1-acid glycoprotein 1 |
| P02774 | GC | Vitamin D-binding protein |
|
| ||
| Neural regulation/growth/development | ||
| Q14118 | DAG1 | Dystroglycan |
| Q16099 | GRIK4 | Glutamate receptor ionotropic, kainate 4 |
| P05067 | APP | Amyloid beta A4 protein |
| A6NNA5 | DRGX | Dorsal root ganglia homeobox protein |
| P36955 | SERPINF1 | Pigment epithelium-derived factor |
| P04053 | DNTT | DNA nucleotidylexotransferase |
| P81605 | DCD | Dermcidin |
| Q7Z5J4 | RAI1 | Retinoic acid-induced protein 1 |
| P55290 | CDH13 | Cadherin-13 |
Figure 8.
Function annotation of the identified secreted proteins. Proteins that have the function annotations in Swiss-Prot database were classified into 4 groups named angiogenesis related proteins, related to neurotrophy, related to cell growth, migration, differentiation, and apoptosis, and proteins associated with inflammation.
4. Discussion
OM-MSCs have significantly clonogenic activity and could be easily propagated for the purpose of transplantation [16]. It indicated that these cells might be obtained from patients themselves and used for autologous transplantation [17, 19–22]. Previous studies have shown that OM-MSCs possessed multidirectional differentiation capacity [16]. As we confirmed here, OM-MSCs have the abilities to differentiate into mesoderm and ectoderm cell type, not only adipose cell or bone cells, but also neurons.
It has been agreed that the secretion of paracrine factors could contribute to the reparative effects of MSCs [25, 32, 33]. So far, the secretion proteome of several MSCs has been investigated [34–38]. However, the secretion proteome of OM-MSCs has not been investigated yet. In the present study we assessed the secretion proteome of MSCs by LC-MS. Our study identified 274 proteins in OM-MSCs conditioned medium and represents to date the first list of secretome of OM-MSCs. The current data showed that about 94.5% of identified proteins were hydrophilic and 63.4% of candidate proteins were located extracellularly. At the same time, it is still found that intracellular proteins from both the cytoplasmic and nuclear compartments accounted for 29% and 22% of candidate proteins. These intracellular proteins may be derived from the death cells.
To enhance our understanding of the paracrine effects of MSCs, the secreted proteins were classified according to their biological processes and molecular functions. Computational analysis predicted many processes that are generally associated with the functions of transplantation such as biological regulation, cellular process, development process, metabolic process, and response to stimulus. These processes would have reparative effects on most injured or diseased tissues, as their facilitation of immune cell migration to the site of injury, ECM remodeling, and an increase in the cellular metabolism [39].
It is believed that the promotion of OM-MSCs transplantation for treating CNS injuries is involved in the following four areas. (1) Cell growth and migration: the genes implicated in cell cycle were present in the OM-MSCs. Transferrin, IGF binding protein family members (IGFBP2, IGFBP3, IGFBP4, and IGFBP7), and SPARC are all reported to be involved in cell proliferation, migration, and differentiation. Plasminogen, microtubule-associated protein, clusterin, and collagen alpha-2 are known to be involved in cell apoptosis. (2) Angiogenesis and blood circulation: OM-MSCs expressed a number of genes implicated in regulation of blood circulation, that is, platelet glycoprotein V, carboxypeptidase N subunit 2, hemopexin, and even regulated angiogenesis, that is, plasminogen, histidine-rich glycoprotein, and pigment epithelium-derived factor (PEDF). (3) Inflammation/immune regulation: many of the genes are implicated in inflammation and immune regulations, that is, complement C3 and C4, CD5 antigen-like, and attractin. (4) Neurotrophy: OM-MSCs also expressed the essential genes that provide nutrition for nerve. Dystroglycan, dorsal root ganglia homeobox protein, and PEDF are known to play key roles in neural regulation, growth, and development. Thus we focused on proteins in these four areas. 45 proteins identified in this work were related in cell cycle like cell growth, development, differentiation, and apoptosis. 27 proteins were associated with the blood cycle, such as angiogenesis and blood coagulation. The immune regulation and neurotrophy related proteins were 28 and 9. Interestingly, there were plenty of proteins that are involved in more than one category especially some of which were tightly associated with neural growth and development suggested that OM-MSCs could be an effective source for repair of injured CNS and may play a critical role in OM-MSCs transplantation.
The IGF is a classical protein family regulating cell growth, survival, and differentiation, which not only is involved in the regulation of brain growth, development, and myelination, but also affects cognition and biochemistry in the adult brain [40, 41]. The IGFBPs have been proposed (1) to act as transport proteins in plasma, (2) to prolong the half-lives of the IGFs in circulation, (3) to determine the tissue- and cell-specific localization of IGF-I and IGF-II, and (4) to control the biological actions of IGF-I and IGF-II by modulating their interactions with their receptors [42]. One of the members is IGFBP-2, which plays a crucial role in regulating cell proliferation, driving invasion, and suppressing apoptosis [43]. Additionally, IGFBP-2 has been demonstrated to associate with the cell membranes of the rat olfactory bulb via proteoglycans in a RGD motif-independent manner [44]. Furthermore, OM-MSCs secrete other IGFs just like IGFBP-3, IGFBP-4, IGFBP-6, and IGFBP-7.
OM-MSCs also secreted molecules involved in neural differentiation and other functions. Dystroglycan can organize axon guidance cue location which is critical for nervous system development [45]. As an ECM, dystroglycan plays important roles in perisynaptic and axonal matrix formations and contributes to synaptic homeostatic plasticity [46]. The loss or decreased expression of dystroglycan could lead to poor prognosis in different malignant tumors [47–49] which suggested that dystroglycan may play the crucial roles during the repair of damage. Other proteins like dermcidin (DCD), retinoic acid induced 1 (RAI1), and cadherin 13 not only contribute to cell cycle related events, but also play roles in neural differentiation. DCD is a neural growth and survival factor and its therapeutic inhibition may be an effective treatment in a subset of breast carcinomas [50]. RAI1 is involved in neurobehavioral disorders [51] and plays a critical role in normal neural and craniofacial development [52]. Cadherin 13 is the member of a family of calcium-dependent cell-cell adhesion proteins and acts as a regulator of neural cell growth [53].
CD248 or endosialin was originally described as tumor endothelial marker which played a role in tumor angiogenesis [54–56]. CD248 is expressed not only in different kinds of tumors or cancer such as human carcinomas, sarcomas, and neuroectodermal tumors [57–59], but also during physiological processes including corpus luteum formation and wound healing [60]. Interestingly, recent studies reveal that the induction of CD248 expression by hypoxia is mediated by HIF-2α [61]. It turned out that CD248 is a marker for activated mesenchymal cells [62]. Therefore, we believe that CD248 could be developed as potential marker of OM-MSCs. Other CD molecules like CD5L may play a role in the regulation of the immune system [63].
Pigment-epithelium-derived factor (PEDF) belongs to serine protease inhibitor (serpin) superfamily that has been widely expressed among many tissues. Traditionally, PEDF is initially identified as a strong antiangiogenic factor [64], while recently increasing evidences demonstrate that PEDF is also a neurotrophic factor [65]. PEDF can improve neuronal survival and protect neurons including motor neurons [66], hippocampal neurons [67], dopaminergic midbrain neurons [65], and striatal neurons [68] in different toxin-induced models. Moreover PEDF has been linked with stem cell biology, and there are now accumulating researches showing PEDF support stem cell survival and maintaining multipotency of stem cells [69].
Besides, we found that the apolipoprotein E, insulin-like growth factor-binding protein 2 (IGF-BP2), clusterin, and other 38 proteins are not only detected in OM-MSC conditioned medium, but also detected in secretion proteome of OECs [70], indicating that there are some certain connections between them.
The paracrine hypothesis of MSCs avoids the immune compatibility, tumorigenicity, infections, and costs caused by the cell-based therapy, so that this will introduce a radically different dimension in regenerative medicine. Proteomics is an involved subject which focuses on large-scale study of proteins and their functions [71]. Through this method, we can discover truly effective components. Our results are valuable in the elucidation of the underlying mechanisms of OM-MSC. Such an approach will have a greater potential for the development of “off-the-shelf” MSC-based therapeutics.
5. Conclusion
Our findings suggested that OM-MSCs could secrete multiple trophic factors which might play important role in CNS regeneration. Although not fully understood, the role of OM-MSCs in neuronal regeneration and endogenous CNS repair functions is now largely acknowledged. Our ongoing study will focus on the roles of these molecules during development and regeneration of the primary olfactory system. It might improve our understanding about OM-MSCs and illuminate the application of OM-MSCs therapeutically.
Supplementary Material
Proteins identified in three parallel tests by mass spectrum were shown in the supplementary table. As the result, a total of 274 nonredundant peoteins in OM-MSC conditioned medium were included in the table.
Acknowledgments
The authors would like to thank Professors Songping Liang, Ying Wang, and Zhonghua Liu from Key Laboratory of Protein Chemistry in Hunan Normal University for their valuable help. This work was supported by National Natural Science Foundation of China (no. 31370817, no. 81070353, and no. 81371358), Hunan Provincial Natural Science Foundation of China (no. 14JJ2060), the Cooperative Innovation Center of Engineering and New Products for Developmental Biology of Hunan Province (no. 20134486), and Scientific Research Fund of Hunan Provincial Education Department (no. 12C0238), Hunan Provincial Innovation Foundation for Postgraduate (no. CX2014B237 and no. CX2015B188).
Conflict of Interests
The authors declare no funding or conflict of interests.
Authors' Contribution
Lite Ge and Miao Jiang contributed equally to this work.
References
- 1.Maguire E. A., Gadian D. G., Johnsrude I. S., et al. Navigation-related structural change in the hippocampi of taxi drivers. Proceedings of the National Academy of Sciences of the United States of America. 2000;97(8):4398–4403. doi: 10.1073/pnas.070039597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reubinoff B. E., Itsykson P., Turetsky T., et al. Neural progenitors from human embryonic stem cells. Nature Biotechnology. 2001;19(12):1134–1140. doi: 10.1038/nbt1201-1134. [DOI] [PubMed] [Google Scholar]
- 3.Yu Y., He J., Zhang Y., et al. Increased hippocampal neurogenesis in the progressive stage of Alzheimer's disease phenotype in an APP/PS1 double transgenic mouse model. Hippocampus. 2009;19(12):1247–1253. doi: 10.1002/hipo.20587. [DOI] [PubMed] [Google Scholar]
- 4.Jin K., Peel A. L., Mao X. O., et al. Increased hippocampal neurogenesis in Alzheimer's disease. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(1):343–347. doi: 10.1073/pnas.2634794100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li B., Yamamori H., Tatebayashi Y., et al. Failure of neuronal maturation in Alzheimer disease dentate gyrus. Journal of Neuropathology & Experimental Neurology. 2008;67(1):78–84. doi: 10.1097/nen.0b013e318160c5db. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lindvall O., Kokaia Z. Stem cells for the treatment of neurological disorders. Nature. 2006;441(7097):1094–1096. doi: 10.1038/nature04960. [DOI] [PubMed] [Google Scholar]
- 7.Aggarwal S., Pittenger M. F. Human mesenchymal stem cells modulate allogeneic immune cell responses. Blood. 2005;105(4):1815–1822. doi: 10.1182/blood-2004-04-1559. [DOI] [PubMed] [Google Scholar]
- 8.Hayashi T., Wakao S., Kitada M., et al. Autologous mesenchymal stem cell-derived dopaminergic neurons function in parkinsonian macaques. The Journal of Clinical Investigation. 2013;123(1):272–284. doi: 10.1172/jci62516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barry F. P., Murphy J. M. Mesenchymal stem cells: clinical applications and biological characterization. The International Journal of Biochemistry & Cell Biology. 2004;36(4):568–584. doi: 10.1016/j.biocel.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 10.Amos T. A. S., Gordon M. Y. Sources of human hematopoietic stem cells for transplantation—a review. Cell Transplantation. 1995;4(6):547–569. doi: 10.1016/0963-6897(95)00037-x. [DOI] [PubMed] [Google Scholar]
- 11.Mueller S. M., Glowacki J. Age-related decline in the osteogenic potential of human bone marrow cells cultured in three-dimensional collagen sponges. Journal of Cellular Biochemistry. 2001;82(4):583–590. doi: 10.1002/jcb.1174. [DOI] [PubMed] [Google Scholar]
- 12.Stenderup K., Justesen J., Clausen C., Kassem M. Aging is associated with decreased maximal life span and accelerated senescence of bone marrow stromal cells. Bone. 2003;33(6):919–926. doi: 10.1016/j.bone.2003.07.005. [DOI] [PubMed] [Google Scholar]
- 13.Murrell W., Féron F., Wetzig A., et al. Multipotent stem cells from adult olfactory mucosa. Developmental Dynamics. 2005;233(2):496–515. doi: 10.1002/dvdy.20360. [DOI] [PubMed] [Google Scholar]
- 14.Delorme B., Nivet E., Gaillard J., et al. The human nose harbors a niche of olfactory ectomesenchymal stem cells displaying neurogenic and osteogenic properties. Stem Cells and Development. 2010;19(6):853–866. doi: 10.1089/scd.2009.0267. [DOI] [PubMed] [Google Scholar]
- 15.Boone N., Bergon A., Loriod B., et al. Genome-wide analysis of familial dysautonomia and kinetin target genes with patient olfactory ecto-mesenchymal stem cells. Human Mutation. 2012;33(3):530–540. doi: 10.1002/humu.22010. [DOI] [PubMed] [Google Scholar]
- 16.Guo Z., Draheim K., Lyle S. Isolation and culture of adult epithelial stem cells from human skin. Journal of Visualized Experiments. 2011;(49, article 2561) doi: 10.3791/2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McDonald C., Mackay-Sim A., Crane D., Murrell W. Could cells from your nose fix your heart? Transplantation of olfactory stem cells in a rat model of cardiac infarction. TheScientificWorldJOURNAL. 2010;10:422–433. doi: 10.1100/tsw.2010.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Toft A., Tomé M., Lindsay S. L., Barnett S. C., Riddell J. S. Transplant-mediated repair properties of rat olfactory mucosal OM-I and OM-II sphere-forming cells. Journal of Neuroscience Research. 2012;90(3):619–631. doi: 10.1002/jnr.22789. [DOI] [PubMed] [Google Scholar]
- 19.Xiao M., Klueber K. M., Zhou J., et al. Human adult olfactory neural progenitors promote axotomized rubrospinal tract axonal reinnervation and locomotor recovery. Neurobiology of Disease. 2007;26(2):363–374. doi: 10.1016/j.nbd.2007.01.012. [DOI] [PubMed] [Google Scholar]
- 20.Xiao M., Klueber K. M., Lu C., et al. Human adult olfactory neural progenitors rescue axotomized rodent rubrospinal neurons and promote functional recovery. Experimental Neurology. 2005;194(1):12–30. doi: 10.1016/j.expneurol.2005.01.021. [DOI] [PubMed] [Google Scholar]
- 21.Nivet E., Vignes M., Girard S. D., et al. Engraftment of human nasal olfactory stem cells restores neuroplasticity in mice with hippocampal lesions. The Journal of Clinical Investigation. 2011;121(7):2808–2820. doi: 10.1172/jci44489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murrell W., Wetzig A., Donnellan M., et al. Olfactory mucosa is a potential source for autologous stem cell therapy for Parkinson's disease. STEM CELLS. 2008;26(8):2183–2192. doi: 10.1634/stemcells.2008-0074. [DOI] [PubMed] [Google Scholar]
- 23.Pandit S. R., Sullivan J. M., Egger V., Borecki A. A., Oleskevich S. Functional effects of adult human olfactory stem cells on early-onset sensorineural hearing loss. STEM CELLS. 2011;29(4):670–677. doi: 10.1002/stem.609. [DOI] [PubMed] [Google Scholar]
- 24.Bas E., Van De Water T. R., Lumbreras V., et al. Adult human nasal mesenchymal-like stem cells restore cochlear spiral ganglion neurons after experimental lesion. Stem Cells and Development. 2014;23(5):502–514. doi: 10.1089/scd.2013.0274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Caplan A. I., Dennis J. E. Mesenchymal stem cells as trophic mediators. Journal of Cellular Biochemistry. 2006;98(5):1076–1084. doi: 10.1002/jcb.20886. [DOI] [PubMed] [Google Scholar]
- 26.Makridakis M., Roubelakis M. G., Vlahou A. Stem cells: insights into the secretome. Biochimica et Biophysica Acta (BBA)—Proteins and Proteomics. 2013;1834(11):2380–2384. doi: 10.1016/j.bbapap.2013.01.032. [DOI] [PubMed] [Google Scholar]
- 27.Salgado A. J. B. O. G., Reis R. L. G., Sousa N. J. C., Gimble J. M. Adipose tissue derived stem cells secretome: soluble factors and their roles in regenerative medicine. Current Stem Cell Research & Therapy. 2010;5(2):103–110. doi: 10.2174/157488810791268564. [DOI] [PubMed] [Google Scholar]
- 28.Hwang S. H., Park S. H., Choi J., et al. Age-related characteristics of multipotent human nasal inferior turbinate-derived mesenchymal stem cells. PLoS ONE. 2013;8(9) doi: 10.1371/journal.pone.0074330.e74330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scintu F., Reali C., Pillai R., et al. Differentiation of human bone marrow stem cells into cells with a neural phenotype: diverse effects of two specific treatments. BMC Neuroscience. 2006;7, article 14 doi: 10.1186/1471-2202-7-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee M. J., Kim J., Kim M. Y., et al. Proteomic analysis of tumor necrosis factor-α-induced secretome of human adipose tissue-derived mesenchymal stem cells. Journal of Proteome Research. 2010;9(4):1754–1762. doi: 10.1021/pr900898n. [DOI] [PubMed] [Google Scholar]
- 31.Dowell J. A., Johnson J. A., Li L. Identification of astrocyte secreted proteins with a combination of shotgun proteomics and bioinformatics. Journal of Proteome Research. 2009;8(8):4135–4143. doi: 10.1021/pr900248y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin R.-Z., Moreno-Luna R., Li D., Jaminet S.-C., Greene A. K., Melero-Martin J. M. Human endothelial colony-forming cells serve as trophic mediators for mesenchymal stem cell engraftment via paracrine signaling. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(28):10137–10142. doi: 10.1073/pnas.1405388111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu C.-H., Hwang S.-M. Cytokine interactions in mesenchymal stem cells from cord blood. Cytokine. 2005;32(6):270–279. doi: 10.1016/j.cyto.2005.11.003. [DOI] [PubMed] [Google Scholar]
- 34.Kwon H. M., Hur S.-M., Park K.-Y., et al. Multiple paracrine factors secreted by mesenchymal stem cells contribute to angiogenesis. Vascular Pharmacology. 2014;63(1):19–28. doi: 10.1016/j.vph.2014.06.004. [DOI] [PubMed] [Google Scholar]
- 35.Xing L., Cui R., Peng L., et al. Mesenchymal stem cells, not conditioned medium, contribute to kidney repair after ischemia-reperfusion injury. Stem Cell Research & Therapy. 2014;5(4, article 101) doi: 10.1186/scrt489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang C.-K., Lee S. K., Luo J., Wang R. H., Dang Q., Chang C. Animal Models for Stem Cell Therapy. Vol. 1213. New York, NY, USA: Springer; 2014. A mouse model of liver injury to evaluate paracrine and endocrine effects of bone marrow mesenchymal stem cells; pp. 69–79. (Methods in Molecular Biology). [DOI] [PubMed] [Google Scholar]
- 37.Guo H.-D., Cui G.-H., Tian J.-X., et al. Transplantation of salvianolic acid B pretreated mesenchymal stem cells improves cardiac function in rats with myocardial infarction through angiogenesis and paracrine mechanisms. International Journal of Cardiology. 2014;177(2):538–542. doi: 10.1016/j.ijcard.2014.08.104. [DOI] [PubMed] [Google Scholar]
- 38.Menezes K., Nascimento M. A., Gonçalves J. P., et al. Human mesenchymal cells from adipose tissue deposit laminin and promote regeneration of injured spinal cord in rats. PLoS ONE. 2014;9(5) doi: 10.1371/journal.pone.0096020.e96020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sze S. K., de Kleijn D. P. V., Lai R. C., et al. Elucidating the secretion proteome of human embryonic stem cell-derived mesenchymal stem cells. Molecular & Cellular Proteomics. 2007;6(10):1680–1689. doi: 10.1074/mcp.m600393-mcp200. [DOI] [PubMed] [Google Scholar]
- 40.Chesik D., Kühl N. M., Wilczak N., De Keyser J. Enhanced production and proteolytic degradation of insulin-like growth factor binding protein-2 in proliferating rat astrocytes. Journal of Neuroscience Research. 2004;77(3):354–362. doi: 10.1002/jnr.20172. [DOI] [PubMed] [Google Scholar]
- 41.Åberg N. D., Brywe K. G., Isgaard J. Aspects of growth hormone and insulin-like growth factor-I related to neuroprotection, regeneration, and functional plasticity in the adult brain. TheScientificWorldJOURNAL. 2006;6:53–80. doi: 10.1100/tsw.2006.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jones J. I., Clemmons D. R. Insulin-like growth factors and their binding proteins: biological actions. Endocrine Reviews. 1995;16(1):3–34. doi: 10.1210/edrv-16-1-3. [DOI] [PubMed] [Google Scholar]
- 43.Fukushima T., Kataoka H. Roles of insulin-like growth factor binding protein-2 (IGFBP-2) in glioblastoma. Anticancer Research. 2007;27(6):3685–3692. [PubMed] [Google Scholar]
- 44.Russo V. C., Bach L. A., Fosang A. J., Baker N. L., Werther G. A. Insulin-like growth factor binding protein-2 binds to cell surface proteoglycans in the rat brain olfactory bulb. Endocrinology. 1997;138(11):4858–4867. doi: 10.1210/en.138.11.4858. [DOI] [PubMed] [Google Scholar]
- 45.Wright K. M., Lyon K. A., Leung H., Leahy D. J., Ma L., Ginty D. D. Dystroglycan organizes axon guidance cue localization and axonal pathfinding. Neuron. 2012;76(5):931–944. doi: 10.1016/j.neuron.2012.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Frischknecht R., Chang K.-J., Rasband M. N., Seidenbecher C. I. Neural ECM molecules in axonal and synaptic homeostatic plasticity. Progress in Brain Research. 2014;214:81–100. doi: 10.1016/B978-0-444-63486-3.00004-9. [DOI] [PubMed] [Google Scholar]
- 47.Sgambato A., Camerini A., Amoroso D., et al. Expression of dystroglycan correlates with tumor grade and predicts survival in renal cell carcinoma. Cancer Biology and Therapy. 2007;6(12):1840–1846. doi: 10.4161/cbt.6.12.4983. [DOI] [PubMed] [Google Scholar]
- 48.Sgambato A., Camerini A., Genovese G., et al. Loss of nuclear p27kip1 and α-dystroglycan is a frequent event and is a strong predictor of poor outcome in renal cell carcinoma. Cancer Science. 2010;101(9):2080–2086. doi: 10.1111/j.1349-7006.2010.01644.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shen J. G., Xu C. Y., Li X., et al. Dystroglycan is associated with tumor progression and patient survival in gastric cancer. Pathology and Oncology Research. 2012;18(1):79–84. doi: 10.1007/s12253-011-9419-2. [DOI] [PubMed] [Google Scholar]
- 50.Porter D., Weremowicz S., Chin K., et al. A neural survival factor is a candidate oncogene in breast cancer. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(19):10931–10936. doi: 10.1073/pnas.1932980100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Carmona-Mora P., Walz K. Retinoic acid induced 1, RAI1: a dosage sensitive gene related to neurobehavioral alterations including autistic behavior. Current Genomics. 2010;11(8):607–617. doi: 10.2174/138920210793360952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tahir R., Kennedy A., Elsea S. H., Dickinson A. J. Retinoic acid induced-1 (Rai1) regulates craniofacial and brain development in Xenopus . Mechanisms of Development. 2014;133:91–104. doi: 10.1016/j.mod.2014.05.004. [DOI] [PubMed] [Google Scholar]
- 53.Patel S. D., Chen C. P., Bahna F., Honig B., Shapiro L. Cadherin-mediated cell-cell adhesion: sticking together as a family. Current Opinion in Structural Biology. 2003;13(6):690–698. doi: 10.1016/j.sbi.2003.10.007. [DOI] [PubMed] [Google Scholar]
- 54.Rettig W. J., Garin-Chesa P., Healey J. H., Su S. L., Jaffe E. A., Old L. J. Identification of endosialin, a cell surface glycoprotein of vascular endothelial cells in human cancer. Proceedings of the National Academy of Sciences of the United States of America. 1992;89(22):10832–10836. doi: 10.1073/pnas.89.22.10832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.St Croix B., Rago C., Velculescu V., et al. Genes expressed in human tumor endothelium. Science. 2000;289(5482):1197–1202. doi: 10.1126/science.289.5482.1197. [DOI] [PubMed] [Google Scholar]
- 56.Seaman S., Stevens J., Yang M. Y., Logsdon D., Graff-Cherry C., St Croix B. Genes that distinguish physiological and pathological angiogenesis. Cancer Cell. 2007;11(6):539–554. doi: 10.1016/j.ccr.2007.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Davies G., Cunnick G. H., Mansel R. E., Mason M. D., Jiang W. G. Levels of expression of endothelial markers specific to tumour-associated endothelial cells and their correlation with prognosis in patients with breast cancer. Clinical & Experimental Metastasis. 2004;21(1):31–37. doi: 10.1023/b:clin.0000017168.83616.d0. [DOI] [PubMed] [Google Scholar]
- 58.Madden S. L., Cook B. P., Nacht M., et al. Vascular gene expression in nonneoplastic and malignant brain. The American Journal of Pathology. 2004;165(2):601–608. doi: 10.1016/s0002-9440(10)63324-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Brady J., Neal J., Sadakar N., Gasque P. Human endosialin (tumor endothelial marker 1) is abundantly expressed in highly malignant and invasive brain tumors. Journal of Neuropathology and Experimental Neurology. 2004;63(12):1274–1283. doi: 10.1093/jnen/63.12.1274. [DOI] [PubMed] [Google Scholar]
- 60.Carson-Walter E. B., Watkins D. N., Nanda A., Vogelstein B., Kinzler K. W., St Croix B. Cell surface tumor endothelial markers are conserved in mice and humans. Cancer Research. 2001;61(18):6649–6655. [PubMed] [Google Scholar]
- 61.Ohradanova A., Gradin K., Barathova M., et al. Hypoxia upregulates expression of human endosialin gene via hypoxia-inducible factor 2. British Journal of Cancer. 2008;99(8):1348–1356. doi: 10.1038/sj.bjc.6604685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bagley R. G., Honma N., Weber W., et al. Endosialin/TEM 1/CD248 is a pericyte marker of embryonic and tumor neovascularization. Microvascular Research. 2008;76(3):180–188. doi: 10.1016/j.mvr.2008.07.008. [DOI] [PubMed] [Google Scholar]
- 63.Tissot J. D., Yamashita H., Nakamura K., et al. IgM are associated to Spα (CD5 antigen-like) Electrophoresis. 2002;23(7-8):1203–1206. doi: 10.1002/1522-2683(200204)23:7/8<1203::AID-ELPS1203>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 64.Dawson D. W., Volpert O. V., Gillis P., et al. Pigment epithelium-derived factor: a potent inhibitor of angiogenesis. Science. 1999;285(5425):245–248. doi: 10.1126/science.285.5425.245. [DOI] [PubMed] [Google Scholar]
- 65.Falk T., Zhang S., Sherman S. J. Pigment Epithelium Derived Factor (PEDF) is neuroprotective in two in vitro models of Parkinson's disease. Neuroscience Letters. 2009;458(2):49–52. doi: 10.1016/j.neulet.2009.04.018. [DOI] [PubMed] [Google Scholar]
- 66.Bilak M. M., Corse A. M., Bilak S. R., Lehar M., Tombran-Tink J., Kuncl R. W. Pigment epithelium-derived factor (PEDF) protects motor neurons from chronic glutamate-mediated neurodegeneration. Journal of Neuropathology and Experimental Neurology. 1999;58(7):719–728. doi: 10.1097/00005072-199907000-00006. [DOI] [PubMed] [Google Scholar]
- 67.DeCoster M. A., Schabelman E., Tombran-Tink J., Bazan N. G. Neuroprotection by pigment epithelial-derived factor against glutamate toxicity in developing primary hippocampal neurons. Journal of Neuroscience Research. 1999;56(6):604–610. doi: 10.1002/(SICI)1097-4547(19990615)56:6<604::AID-JNR6>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 68.Sanagi T., Yabe T., Yamada H. Adenoviral gene delivery of pigment epithelium-derived factor protects striatal neurons from quinolinic acid-induced excitotoxicity. Journal of Neuropathology and Experimental Neurology. 2010;69(3):224–233. doi: 10.1097/nen.0b013e3181cfc46f. [DOI] [PubMed] [Google Scholar]
- 69.Elahy M., Baindur-Hudson S., Dass C. R. The emerging role of PEDF in stem cell biology. Journal of Biomedicine and Biotechnology. 2012;2012:6. doi: 10.1155/2012/239091.239091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Liu Y., Teng X., Yang X., et al. Shotgun proteomics and network analysis between plasma membrane and extracellular matrix proteins from rat olfactory ensheathing cells. Cell Transplantation. 2010;19(2):133–146. doi: 10.3727/096368910x492607. [DOI] [PubMed] [Google Scholar]
- 71.Kupcova Skalnikova H. Proteomic techniques for characterisation of mesenchymal stem cell secretome. Biochimie. 2013;95(12):2196–2211. doi: 10.1016/j.biochi.2013.07.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Proteins identified in three parallel tests by mass spectrum were shown in the supplementary table. As the result, a total of 274 nonredundant peoteins in OM-MSC conditioned medium were included in the table.