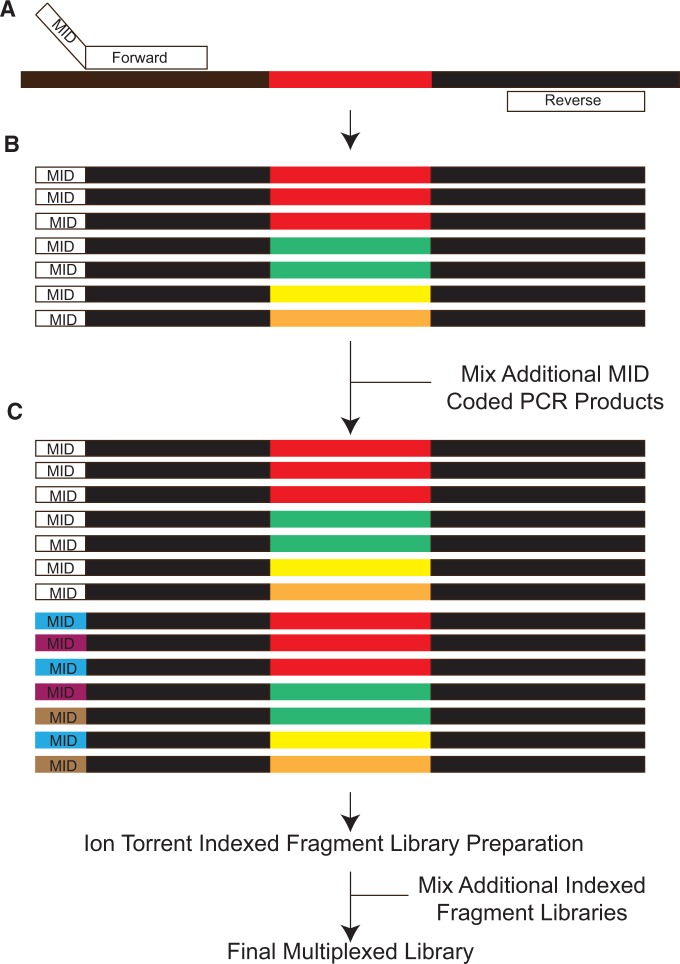
Figure 2.
Schematic of library preparation for highly multiplexed amplicon deep sequencing. (A) Samples are amplified using primers targeting conserved sequences (black fragments) surrounding variable regions (colored fragments). The forward primer contains a barcode (MID) specific to the sample and replicate. (B) After amplification, each PCR reaction contains amplicons representing the haplotypes within that sample labeled with the specific MID. (C) Multiple samples are then mixed forming a final library containing multiple MIDs (white, blue, purple and brown) for preparation for sequencing. This mixture is library prepped with a specific index placed on the library during the process. Multiple libraries with different indexes can then be pooled and sequenced at the same time allowing for highly multiplexed sequencing