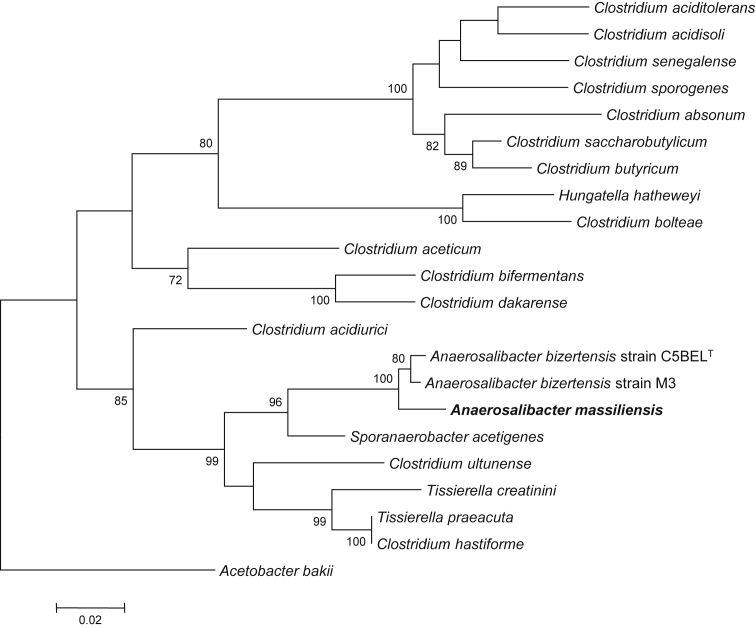
Fig. 1.
Phylogenetic tree highlighting position of Anaerosalibacter massiliensis sp. nov. strain ND1T relative to other type strains within Clostridiaceae. Strains and their corresponding GenBank accession numbers for 16S rRNA genes are (type = T): A. massiliensis strain ND1T, HG315673; A. bizertensis strain C5BELT, HQ534365[3]; A. bizertensis strain M3, HG964477; S. acetigenes strain Lup33T, NR_025151 [28]; C. ultunense strain BST, GQ461825[29]; T. creatinini strain BN11T, FR749955[30]; C. hastiforme strain ATCC 33268T, X80841[23], [31]; T. preacuta strain ATCC 25539T, GQ461814[32]; C. acidurici strain ATCC 7906T, M59084[23], [33]; C. aceticum strain ATCC 35044T, Y18183[34]; C. bifermentans strain ATCC 638T, AB075769[23], [35]; C. dakarense strain FF1T, KC517358[36]; C. saccharobutylicum strain ATCC BAA-117T, U16147[37]; C. butyricum strain ATCC 19398T, AJ458420[23], [38]; C. absonum strain ATCC 27555T, X77842[39]; C. senegalense strain JC122T, NR_125591 [7]; C. sporogenes strain ATCC 3584T, X68189[23], [35]; C. aciditolerans strain JW/YJL-B3T, DQ114945[40]; C. acidisoli strain CK74T, AJ237756[41]; Hungatella hatheweyi strain UB-B.2T, HE603919[42]; C. bolteae strain ATCC BAA-613T, AJ508452[43], [44]. 16S rRNA from A. massiliensis (1512 bp) was amplified and sequenced using fd1 (5′- AGAGTTTGATCCTGGCTCAG-3′) and rP2 (5′-ACGGCTACCTTGTTACGACTT-3′) primers; 16S rRNA sequences from all studied strains were aligned using CLUSTALW; total of 1182 nucleotide positions present in all studied sequences were used for phylogenetic inferences with maximum-likelihood method within MEGA6 software. Numbers at nodes are percentages of bootstrap values obtained by repeating analysis 500 times to generate majority consensus tree. Only bootstrap values greater than 70% are indicated. Acetobacterium bakii strain DSM 8239T, X96960[45], was used as outgroup. Scale bar = 2% nucleotide sequence divergence.