Abstract
Virus assembly is a coordinated process in which typically hundreds of subunits react to form complex, symmetric particles. We use resistive-pulse sensing to characterize the assembly of Hepatitis B Virus core protein dimers into T = 3 and T = 4 icosahedral capsids. This technique counts and sizes intermediates and capsids in real time, with single particle sensitivity, and at biologically relevant concentrations. Other methods are not able to produce comparable real-time, single-particle observations of assembly reactions below, near, and above the pseudocritical dimer concentration, at which the dimer and capsid concentrations are approximately equal. Assembly reactions across a range of dimer concentrations reveal three distinct patterns. At dimer concentrations as low as 50 nM, well below the pseudo-critical dimer concentration of 0.5 µM, we observe a switch in the ratio of T = 3 to T = 4 capsids, which increases with decreasing dimer concentration. Far above the pseudo-critical dimer concentration, kinetically trapped, incomplete T = 4 particles assemble rapidly, then slowly anneal into T = 4 capsids. At all dimer concentrations tested, T = 3 capsids form more rapidly than T = 4 capsids, suggesting distinct pathways for the two forms.
Keywords: nanofluidics, in-plane nanochannel, resistive-pulse sensing, single-particle counting, Hepatitis B Virus, self-assembly
Understanding the general principles of virus self-assembly has potential applications in virology,1 antiviral therapy,2–4 and biotechnology.5–6 For these applications to be realized, we must know the timescales and conditions that impact assembly both constructively and destructively. Many viral capsid proteins can spontaneously assemble in vitro in response to solution conditions, e.g., initial subunit concentration, temperature, pH, and small molecule assembly effectors. A number of these capsid proteins are readily available through various expression systems, which facilitates studying the assembly process. However, we lack experimental methods that can effectively probe the mechanisms of assembly.
Hepatitis B Virus (HBV) makes a tractable model system to study assembly. In vitro capsid assembly requires a single capsid protein and is well characterized from bulk solution experiments.7 Moreover, HBV has a broad impact on human health;8 consequently, the viral core (or capsid) protein is a target of antiviral drug development.7 The best characterized activity of the core protein is its ability, in vivo and in vitro, to self-assemble to form icosahedral capsids from 90 or 120 core protein homodimers. These capsids are arranged with T = 3 (32-nm diameter) or T = 4 (35-nm diameter) icosahedral symmetry, respectively.9 The core protein assembly domain (Cp149), a truncated version of the 183-residue protein that is missing the RNA-binding domain, assembles in vitro as a function of protein concentration, ionic strength, and temperature.10–11 In this study, we examine Cp149 dimers that form empty virus-like particles (or capsids) that are morphologically indistinguishable from capsids isolated from cell cultures.12–14
A nucleation-growth model has been proposed to explain HBV assembly.15–16 The formation of a nucleus, either a specific structure or an irregular nucleating complex,16 is the kinetically limiting step.15,17 The lag phase of the sigmoidal assembly curve represents the formation of a steady state, or shock front,18 of intermediates19–20 and is followed by rapid capsid accumulation until the reaction approaches equilibrium or free subunits are depleted. The combination of a nucleation step and weak subunit-subunit interaction (−3 to −4 kcal/mol for each of the four contacts made by each dimer) leads to a directed assembly that is less susceptible to kinetic traps as compared to non-nucleated assembly.1,21 However, reactions can become kinetically trapped when association energy is very high (e.g., at high salt), at high dimer concentrations, or both, which can lead to too many nuclei and formation of incomplete capsids due to depletion of subunit.15,19
Tracking the changes in dimer, intermediate, and capsid concentrations simultaneously requires measurement of particle sizes with broad dynamic range, high resolution, and temporal precision. Classical and dynamic light scattering measurements are used to extract assembly kinetics of bacteriophage P22,22 Hepatitis B Virus,15 Cowpea Chlorotic Mottle Virus,23–24 Brome Mosaic Virus,25 and Human Papillomavirus,26 but these studies only report a signal proportional to average molecular weight, obscuring the identities of individual components. Similarly, size exclusion chromatography (SEC), often used to quantify assembled capsids,11 lacks the resolution to identify specific intermediates and capsids. Even in small angle X-ray scattering (SAXS) studies, which are limited to high concentrations of solute, the contributions of major species are not easily extracted.27,28 Other methods used to study virus assembly include time-offlight mass spectrometry29 and charge-detection mass spectrometry30 (CD-MS), of which CD-MS is able to detect single particles. Unfortunately, electrospray ionization mass spectrometry is only compatible with volatile buffers (e.g., ammonium acetate), consequently, limiting the selection of assembly conditions. Fluorescence correlation spectroscopy31 counts single particles, but is limited to very low concentrations.
Resistive-pulse sensing (or Coulter counting) measures a change in conductivity as a particle transits an electrically biased nanopore of comparable dimension, and the measurement is sensitive to subtle changes in analyte size. Resistive-pulse sensing has been used to detect a variety of viruses, including HBV capsid particles,32–34 a Paramecium busaria chlorella virus-antibody complex,35 Human Papillomavirus,36 and the filamentous virus fd.37 Resistive-pulse sensing is able to provide thermodynamic and kinetic information, as well as identify intermediate and capsid species. Furthermore, the limit of detection is far below most methods used to study virus assembly, which permits study of assembly well below the pseudo-critical concentration and does not require fluorescent labeling of the dimer.
Here, we use an in-plane nanofluidic device with two pores in series33–34 to investigate assembly of HBV Cp149 dimers into empty virus-like particles in 1 M NaCl under three distinct regimes: (i) assembly below the pseudo-critical dimer concentration, (ii) assembly near the pseudo-critical dimer concentration, typically expected to lead to assembly of normal capsids, and (iii) assembly at high dimer concentrations that favor formation of kinetically trapped intermediates. Assembly near the pseudo-critical concentration, at which the dimer and capsid concentrations are approximately equal, provides a view of assembly that is most easily compared with earlier publications. Below the pseudo-critical concentration, we report HBV assembly for dimer concentrations as low as 50 nM and obtain a new view of assembly kinetics. At high dimer concentration (> 2 µM), incomplete capsids are trapped and slowly anneal into T = 4 capsids.
Results
Resistive-Pulse Sensing of Capsids and Their Intermediates
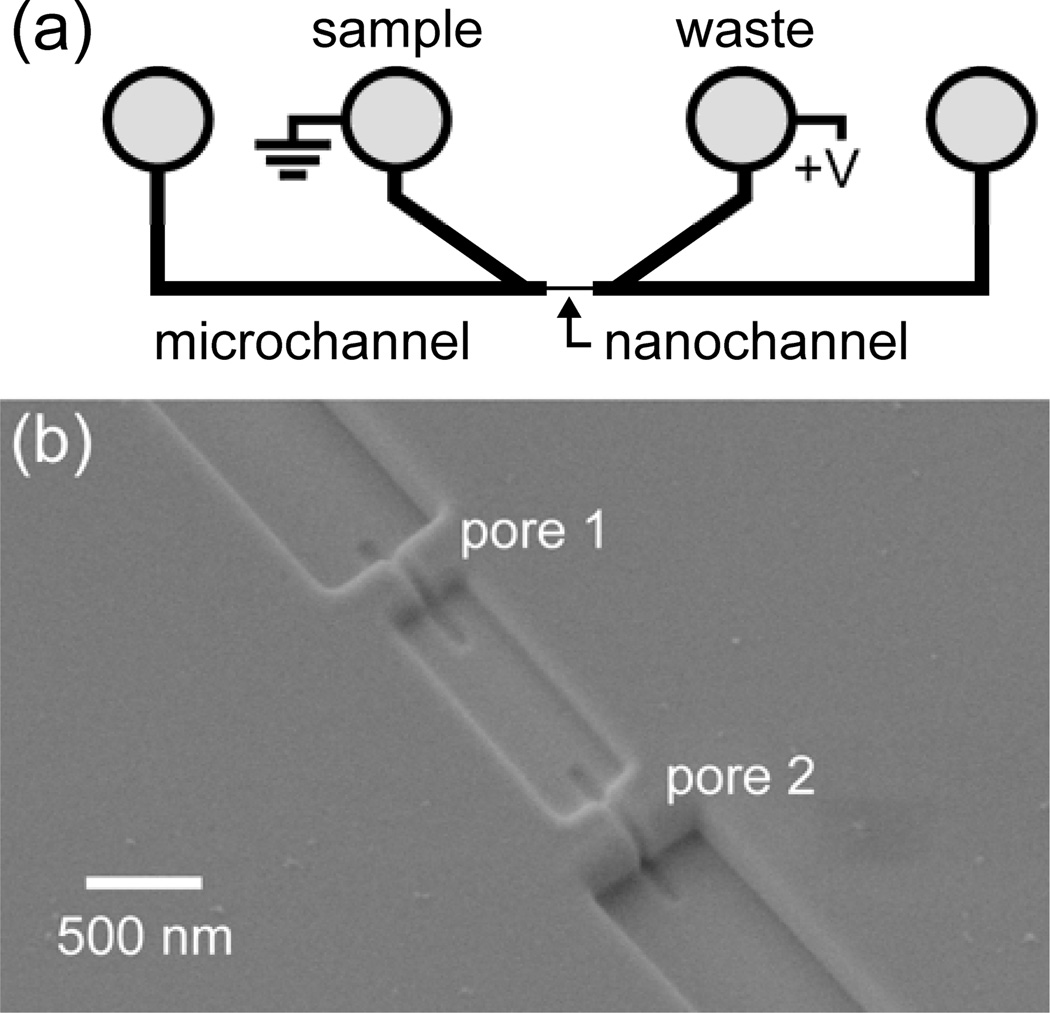
Resistive-pulse measurements were made with the device design shown in Figure 1. Two glass microchannels are bridged by a 10-µm long nanochannel with two nanopores in series (Figure 1b). The nanopores are 200 ± 10 nm long, 60 ± 5 nm wide, and 60 ± 5 nm deep and tailored to sense the 32-nm T = 3 and 35-nm T = 4 HBV capsids, as well as intermediates formed in the reaction. HBV capsids and intermediates are electrokinetically driven from one side of the nanofluidic device to the other side through the bridging nanochannel. During translocation through the two pores, each particle produces two pulses (or decreases in the baseline current) (Figure 2a–b). From the resistive-pulse measurements of the T = 3 capsids, T = 4 capsids, and intermediates, we use the pulse amplitude and frequency to determine particle size and concentration, respectively. The average pulse amplitude from the two pores is normalized by the baseline current to calculate Δi/i (relative pulse amplitude), which is proportional to the volume of electrolyte displaced by the volume of capsid protein in each particle. The relative pulse amplitudes (Δi/i) are plotted as histograms to show the distributions of T = 3 capsids, T = 4 capsids, and intermediates (non-capsid oligomers) present during the assembly reactions (Figure 2c–d). Each device is calibrated with a purified T = 3 and T = 4 capsid standard (Figure S1 in the Supporting Information). Pulse amplitudes from pores 1 and 2 differ by less than 10%, which arises from slight differences in the pore dimensions. However, the average pulse amplitude from the two measurements leads to improved precision compared to single-pore measurements and, thus, enhanced size resolution of capsids and intermediates. Moreover, the unique two-pulse current signature for each particle also provides a means to discriminate real events from false positives.
Figure 1.
(a) Schematic of glass nanofluidic device with two V-shaped microchannels separated by a nanochannel with two pores in series. Hepatitis B Virus (HBV) capsids, dimer, or assembly products in a high-salt buffer are loaded into the sample reservoir and electrokinetically driven through the nanochannel with an applied potential. (b) Scanning electron microscope (SEM) image of the nanochannel with two pores milled with a focused ion beam instrument. The nanopore dimensions are 60-nm wide, 60-nm deep, and 200-nm long, and the nanochannel between pores 1 and 2 is 300-nm wide, 100-nm deep, and 1-µm long. Each particle is sensed twice, once at each nanopore.
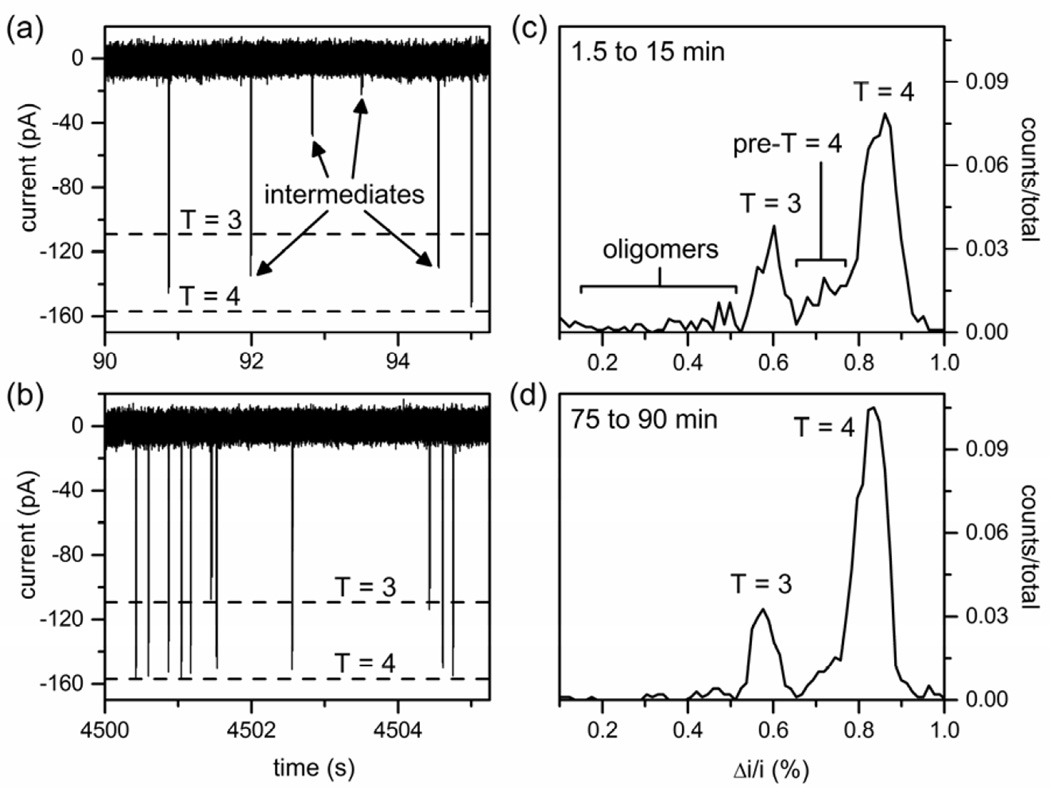
Figure 2.
Variation of current with time from resistive-pulse measurements of particles from an assembly reaction of 0.2 µM Cp149 dimer in 1 M NaCl after (a) 90 s and (b) 75 min. Each particle produces two fully resolved current pulses34 that appear as a single pulse on the timescale used in the figure. Histograms of relative pulse amplitude (Δi/i) of the assembly products for reaction times of (c) 1.5 to 15 min and (d) 75 to 90 min. The Δi/i distributions contain T = 3 capsids, T = 4 capsids, and intermediates (non-capsid oligomers and pre-T = 4 intermediates). Total counts are 1023 and 988 in panels (c) and (d), respectively.
Resistive-pulse measurements of HBV assembly were performed over a range of dimer and salt concentrations. Capsid assembly was initiated by mixing Cp149 dimer (0.05 to 10 µM) with a high salt buffer (0.3 to 1 M NaCl). T = 3 capsids, T = 4 capsids, and intermediates were tracked simultaneously by the resistive-pulse measurements. Assembly with initial dimer concentrations < 2 µM was measured directly by loading the entire reaction mixture onto the nanofluidic device. After a 90 s delay, which corresponded to the time to mix and load the reaction mixture into the device, the assembly reactions were monitored continuously for 90 min. However, for assembly with initial dimer concentrations ≥ 2 µM, the reaction was diluted at various time points from 15 s to 24 h to enable single-particle counting without pulse overlap; dilution also slows down the assembly reaction, which allows intermediate structures to persist for hours. These single-particle measurements are robust and highly reproducible. This manuscript contains data from 54 independent assembly reactions measured on six nanofluidic devices. Detection of > 700,000 virus particles is reported.
Figure 2c–d shows histograms of Δi/i for the assembly products formed from 0.2 µM dimer in 1 M NaCl over a 90 min reaction. At early times, the assembly reactions produce a wide variety of intermediate structures that include non-capsid oligomers smaller in size than T = 3 capsids and pre-T = 4 intermediates between the sizes of T = 3 and T = 4 capsids (Figure 2a and 2c). As the reaction proceeds, fewer and fewer of these intermediates are detected (Figure 2b and 2d). T = 3 and T = 4 capsids are the primary particles present and are easily differentiated as they displace 0.58% and 0.83% of the baseline current (Δi/i), respectively. Intermediates are present with notable Δi/i distributions at 0.71% (105 dimers) and 0.47% (77 dimers). However, almost all of these intermediates are recorded in the first few minutes of the assembly reaction.
The frequency of particle translocation events in counts/min is directly proportional to the particle concentration in solution (Figure S2 in the Supporting Information). From the pulse frequency data, we determine the relative fractions of T = 3 capsids, T = 4 capsids, and intermediates formed to reveal the time and concentration dependence of assembly. The fraction of each species (F) formed per unit time is calculated from Eq. 1 as:
| (1) |
where N is the number of counts in each time interval for T = 3 capsids, T = 4 capsids, and intermediates. Assembly of 0.2 µM dimer in 1 M NaCl produced a capsid distribution of 65% T = 4 capsid and 20% T = 3 capsid, and assembly of 0.9 µM dimer in 1 M NaCl produced 74% T = 4 capsid and 20% T = 3 capsid (Figure S3 in the Supporting Information). Previous studies have shown that assembly of Cp149 dimer in 300 mM NaCl produces 90% to 95% T = 4 capsid and 5% to 10% T = 3 capsid.38
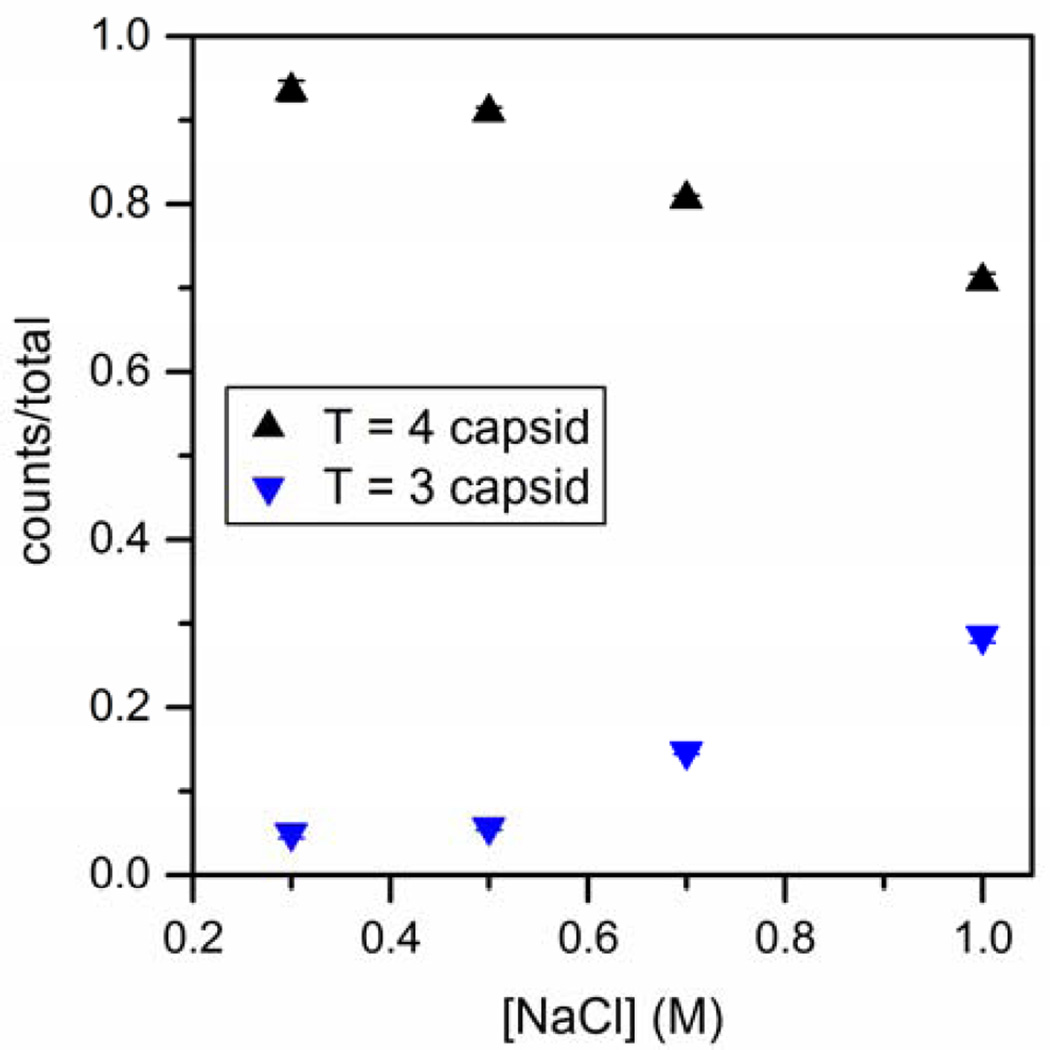
Because we observed a larger fraction of T = 3 capsids than usual, we tested the effect of ionic strength and found that lower NaCl concentration yields fractions of T = 3 and T = 4 capsids similar to those observed in similar conditions in bulk experiments.10,38–39 In Figure 3, the fractions of T = 3 and T = 4 capsids as a function of salt concentration after 90 min of assembly are shown for concentrations of 2 µM dimer in 0.5, 0.7, and 1 M NaCl and 4 µM dimer in 300 mM NaCl. From 0.3 to 1 M NaCl, the fraction of T = 4 capsid decreases. The change in the ratio of T = 3 to T = 4 capsids with salt concentration suggests that NaCl influences dimer conformation or dimer-dimer interactions, altering the preferred geometry of capsid particles. These results also confirm that the high concentrations of T = 3 capsid observed in the assembly experiments were not initially present in the dimer solution, but are instead products of assembly reactions in 1 M NaCl.
Figure 3.
Variation of the fraction of T = 3 and T = 4 capsids with NaCl concentration for 90- min assembly reactions. Initial dimer concentrations are 2 µM for reactions in 0.5, 0.7, and 1 M NaCl and 4.0 µM in 0.3 M NaCl. Products from all reactions are measured by resistive-pulse sensing in 1 M NaCl. Each data point represents three reactions, and error bars are ±σ. Total counts are > 3000 capsids for each salt concentration.
Thermodynamics of HBV Assembly at 1 M NaCl
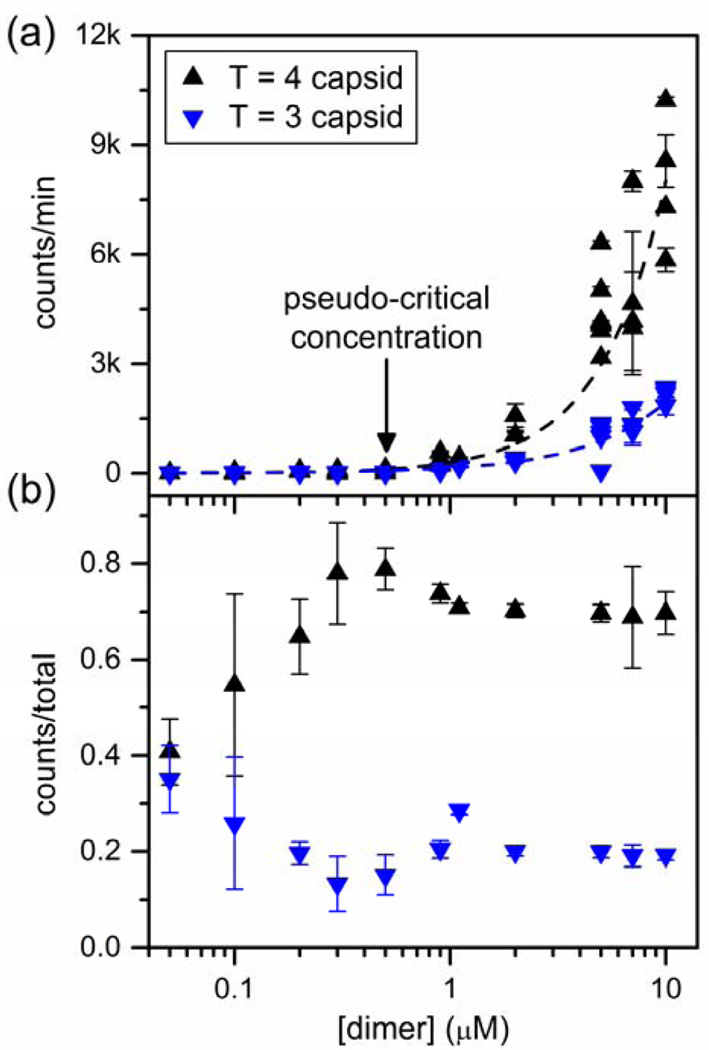
Thermodynamics of capsid assembly are described with good approximation by the law of mass action, and the concentration of dimer at equilibrium is a nearly constant pseudo-critical concentration.19 The counts/min of T = 3 and T = 4 capsids at equilibrium, as a function of initial Cp149 dimer concentration, was determined by resistive-pulse sensing (Figure 4a). A pseudo-critical dimer concentration of 0.5 µM dimer is indicated by the rapid increase in the counts/min, i.e., capsid concentration (Figure S2 in Supporting Information). Resistive-pulse measurements showed that for initial dimer concentrations > 0.5 µM, essentially all additional dimer was found as capsid. However, we observe virus assembly with our nanofluidic devices at concentrations as low as to 0.05 µM dimer, which is consistent with predictions for a pseudo-critical concentration, not a true critical concentration.
Figure 4.
(a) Variation of average counts/min for T = 3 and T = 4 capsids with initial Cp149 dimer concentration in resistive-pulse sensing measurements after a 60-min assembly reaction in 1 M NaCl. The pseudo-critical dimer concentration is 0.5 µM. (b) Variation of the fraction of T = 3 and T = 4 capsids with initial Cp149 dimer concentration in 1 M NaCl. Data in panels (a) and (b) are taken from 54 assembly reactions made on 5 devices and are based on more than 700,000 particles. Error bars are ±σ.
In addition, single-particle counting allowed the contribution from T = 3 and T = 4 capsid to be separated from total particle count. Fractions of T = 3 and T = 4 capsids produced in each assembly reaction are plotted in Figure 4b as a function of dimer concentration from 0.05 to 10 µM. The ratio of T = 3 to T = 4 capsids is constant above the pseudo-critical concentration, consistent with earlier observations.10 Low dimer concentration (0.05 – 0.2 µM) favors formation of T = 3 capsids. As dimer concentration decreases, the fraction of T = 3 capsid progressively increases whereas the fraction of T = 4 capsid produced decreases. At the lowest concentration that assembly was observed (0.05 µM dimer), the relative fractions of T = 3 and T = 4 capsids are nearly identical at 40% each. Near the pseudo-critical concentration (0.5 – 1 µM dimer), the fraction of T = 3 capsids increased slightly with dimer concentration, whereas the fraction of T = 4 capsids decreased. At higher dimer concentrations (2 to 10 µM), there was a slight increase in the ratio of T = 3 to T = 4 with fractions 20% and 70%, respectively. At high dimer concentrations, we hypothesize that this increase in the T = 3 / T = 4 ratio is due to a kinetically trapped pre-T = 4 intermediates.
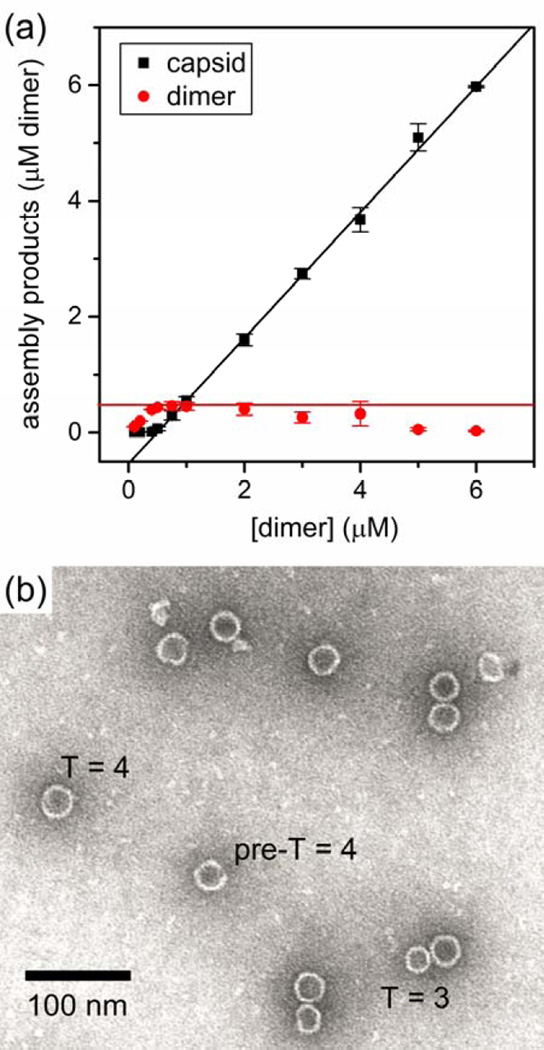
In parallel, we conducted conventional measurements. Assembly reactions of Cp149 dimer were given 24 h to equilibrate, and then concentrations of capsid and dimer were determined by size-exclusion chromatography (SEC) (Figure 5a). Below the pseudo-critical concentration of about 0.5 µM dimer, almost no capsid assembly was observed, whereas above the pseudo-critical concentration, almost all dimer assembled into capsid, leaving a constant concentration of free dimer. Equilibration of the assembly reaction is indicated when the concentration of free dimer can be fitted to a linear regression with a slope of zero.11 However, for initial dimer concentrations ≥ 2 µM, the free dimer concentration appeared to decline at higher initial dimer concentrations, which suggests that a kinetic trap is formed in which large intermediates (i.e., incomplete capsids) were assembled that could not be resolved by SEC. Evidence of large intermediates and capsids missing subunits was confirmed by transmission electron microscopy (TEM) (Figure 5b and Figure S4 in the Supporting Information).
Figure 5.
(a) Variation of the amount of capsid (T = 3 and T = 4) and remaining Cp149 dimer with initial dimer concentration after a 24-h assembly reaction in 1 M NaCl. Amounts of dimer and capsid were determined by size-exclusion chromatography. The pseudo-critical dimer concentration is estimated to be 0.5 µM (red line). Each data point is an average of three assembly reactions, and the error bars are ±σ. (b) Transmission electron microscopy (TEM) image of capsids formed from 5 µM Cp149 dimer after 15 s of reaction in 1 M NaCl. A T = 3 capsid, T = 4 capsid, and incomplete pre-T = 4 intermediate are labeled.
Together, resistive-pulse and SEC data identify the same pseudo-critical dimer concentration. These data allow us to divide capsid assembly at 1 M NaCl into three regimes according to the initial dimer concentration: (i) below the pseudo-critical concentration (< 0.5 µM dimer), (ii) near the pseudo-critical concentration (0.5 to 1 µM dimer), and (iii) substantially above the pseudo-critical concentration (2 to 10 µM dimer). The second regime, near and above the pseudo-critical concentration, is typical for most bulk experiments whereas the third regime, substantially above the pseudo-critical concentration, is characterized by a kinetic trap. In the following sections, resistive-pulse sensing is used to investigate initial dimer concentrations below and above the critical dimer concentration in more detail.
Assembly below the Pseudo-Critical Dimer Concentration
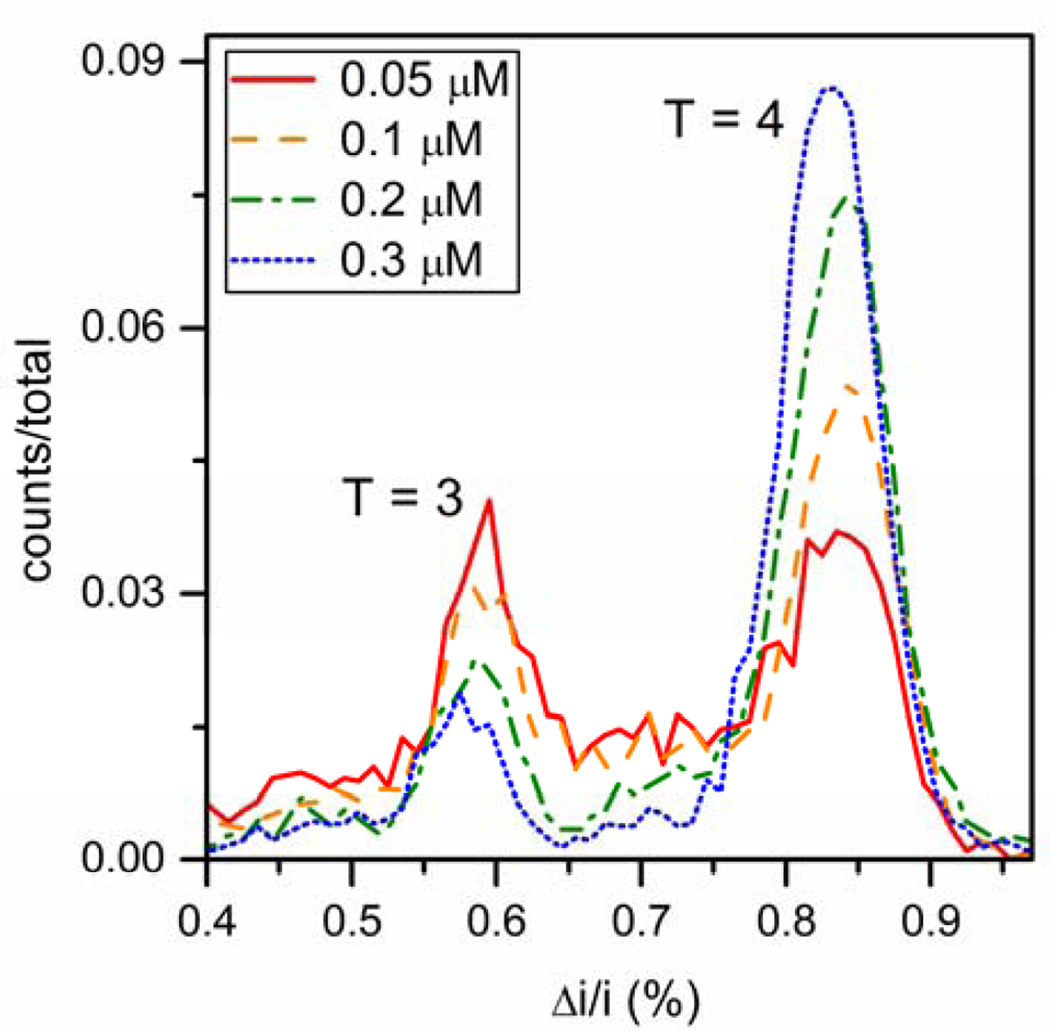
Due to detection limitations of TEM, SEC, and multi-angle laser light scattering (MALLS), little is known about virus assembly below the pseudo-critical dimer concentration (< 0.5 µM dimer in 1 M NaCl). However, the subpseudo- critical concentration range is easily accessed by resistive-pulse sensing. Assembly products for dimer concentrations from 0.05 to 0.3 µM dimer are shown in Figure 6. As the initial dimer concentration decreases, the fraction of T = 4 capsid produced likewise decreases, whereas the fraction of T = 3 capsid increases. At 0.05 µM dimer, the lowest concentration that was tested, the fractions of T = 3 and T = 4 capsids formed were both ~40%.
Figure 6.
Histograms of relative pulse amplitude (Δi/i) of the assembly products formed from 0.05, 0.1, 0.2, and 0.3 µM Cp149 dimer in 1 M NaCl. Below the pseudo-critical dimer concentration (0.5 µM), T = 3 capsid formation is favored. As initial dimer concentration decreases, the amount of T = 4 capsid formed decreases, whereas the amount of T = 3 capsid formed increases. Total counts are > 3000 particles for each dimer concentration.
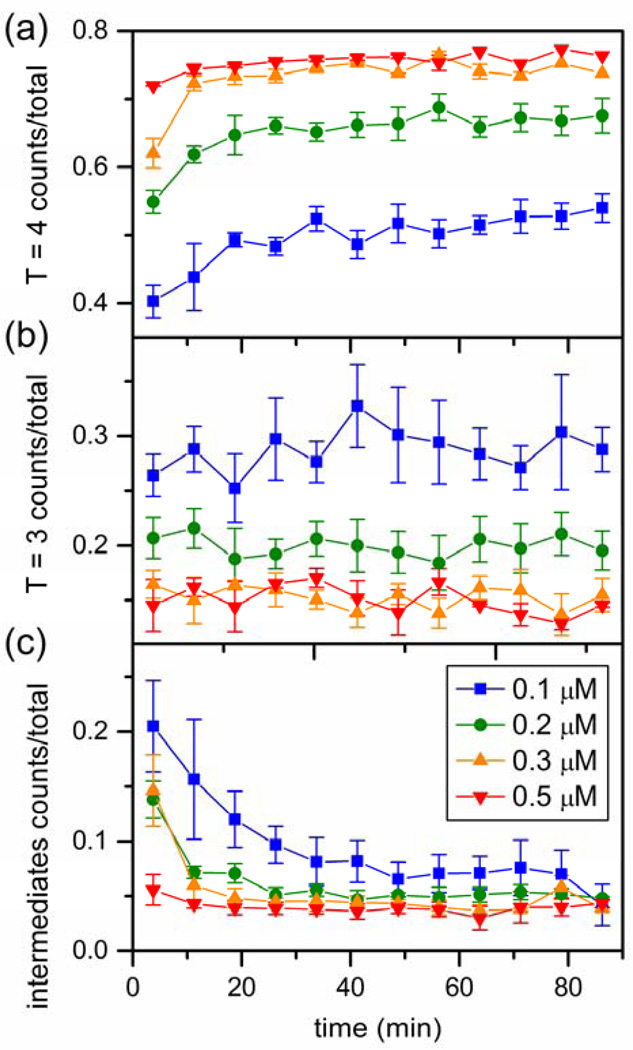
Resistive-pulse measurements have sufficient sensitivity that even at very low concentrations we were able to evaluate time courses for each reaction (Figure 7). Time to achieve a steady state concentration of T = 4 capsid is proportional to initial dimer concentration (Figure 7a). Assembly of T = 4 capsids at 0.1 µM dimer only reaches steady state after 30 min. By comparison, at 0.5 µM initial dimer concentration, T = 4 capsids approach steady state within 5 min. These data allow determining elongation rates of the assembly (i.e., the second order rate that subunits add to a growing nascent capsid). The elongation rate is proportional to the lag time between initiating assembly and the first appearance of capsids.16 Lag times for dimer concentrations from 0.05 to 0.9 µM were extracted from the fraction of T = 4 capsids plotted as a function of time and fitted with Eq. S1 (Figure S5a in the Supporting Information). The slope of the line fitted to the lag time versus the reciprocal of the dimer concentration is proportional to the elongation rate (Eq. S2 and Figure S5b in the Supporting Information). The elongation rate constant is 1.71 × 107 M−1s−1, which is in reasonable agreement with the rate constant of reduced dimer (2.7 × 107 M−1s−1) determined from light scattering data at a lower NaCl concentration but at higher temperature.38
Figure 7.
Variation of the fraction of (a) T = 4 capsid, (b) T = 3 capsid, and (c) intermediates with time for initial Cp149 dimer concentrations from 0.1 to 0.5 µM assembled in 1 M NaCl. In panel (a), the rate of T = 4 capsid growth increases as dimer concentration increases. (b) No change in the T = 3 capsid fraction over time is observed at any dimer concentration. (c) Intermediates include non-capsid oligomers and pre-T = 4 intermediates and decrease over time. Each trajectory is an average of at least three independent assembly reactions, and error bars are ± σ/2. Total counts are > 15,000, 25,000, 23,000, and 27,000 particles for 0.1, 0.2, 0.3, and 0.5 µM dimer, respectively.
The fraction of T = 3 capsid is almost constant over the 90 s to 90 min time interval for all dimer concentrations tested. Apparently, the T = 3 capsid population reaches steady state before the measurement starts and long before the T = 4 capsid population. The intermediates (noncapsid oligomers and pre-T = 4 intermediates) decrease over time; however, low concentrations of these intermediates are present at all times. In terms of mass fraction (Eq. 1), these putative intermediates have an inverse relationship with dimer concentration; consequently, intermediates have the greatest presence at low initial dimer concentration. As the concentration of intermediates decreases, there is a concurrent increase in the T = 4 capsid population, suggesting that these intermediates complete into T = 4 capsids.
Assembly Substantially Above the Pseudo-Critical Concentration
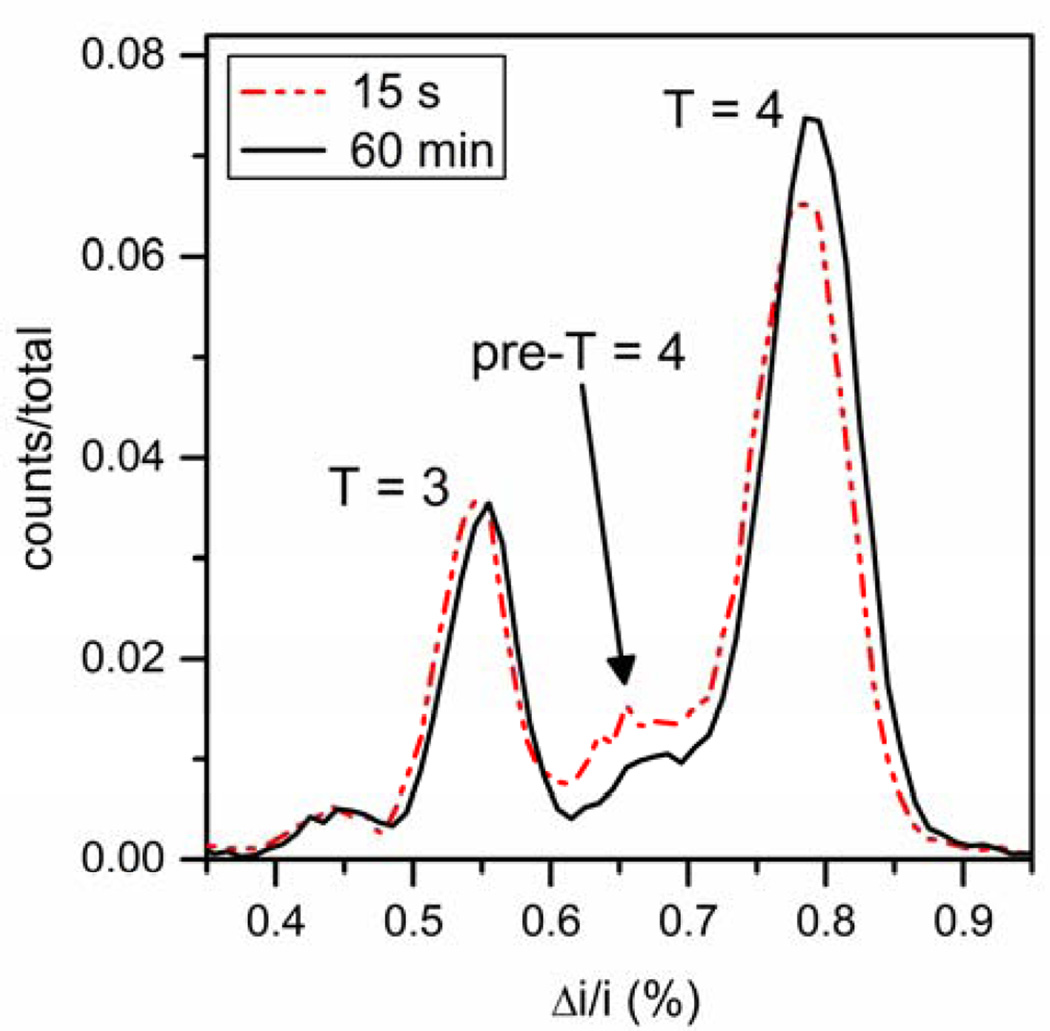
A different population of intermediates is detected above the pseudo-critical dimer concentration (Figure 8). After 15 s of reaction time, a large distribution of pre-T = 4 intermediates with sizes between the sizes of T = 3 and T = 4 capsids was detected. After 60 min, an increase in T = 4 capsids was observed roughly proportional to the amount of these pre-T = 4 intermediates depleted. Intermediates are nearly absent after 24 h. The size of the predominant pre-T = 4 intermediate is estimated by its current displacement to be 105 dimers. Incomplete capsids, presumably intermediates, are also observed by negative stain TEM in reactions quenched at 15 s by blotting onto a TEM grid (Figure 5b and Figure S4 in the Supporting Information). TEM analysis of reactions incubated for 24 h had a smaller proportion of intermediates, in agreement with resistive-pulse measurements. Pre-T = 4 intermediates were detected for dimer concentrations of 5, 7, and 10 µM by both TEM and resistive-pulse sensing. The data in Table S1 in the Supporting Information indicate that the nanochannel neither distorted HBV assembly nor biased detection toward smaller capsids.
Figure 8.
Histograms of relative pulse amplitude (Δi/i) of the assembly products from 5 µM Cp149 dimer in 1 M NaCl after 15 s and 60 min of reaction time. The pre-T = 4 intermediate distribution between the T = 3 and T = 4 capsid distributions has a maximum corresponding to 105-dimer species and decreases in height over time. Little to no change is observed in the T = 3 capsid distribution, and the T = 4 capsid distribution increases in height, which suggests that the pre-T = 4 intermediate peak anneals to form T = 4 capsids. Total counts are > 37,000 particles for each reaction time.
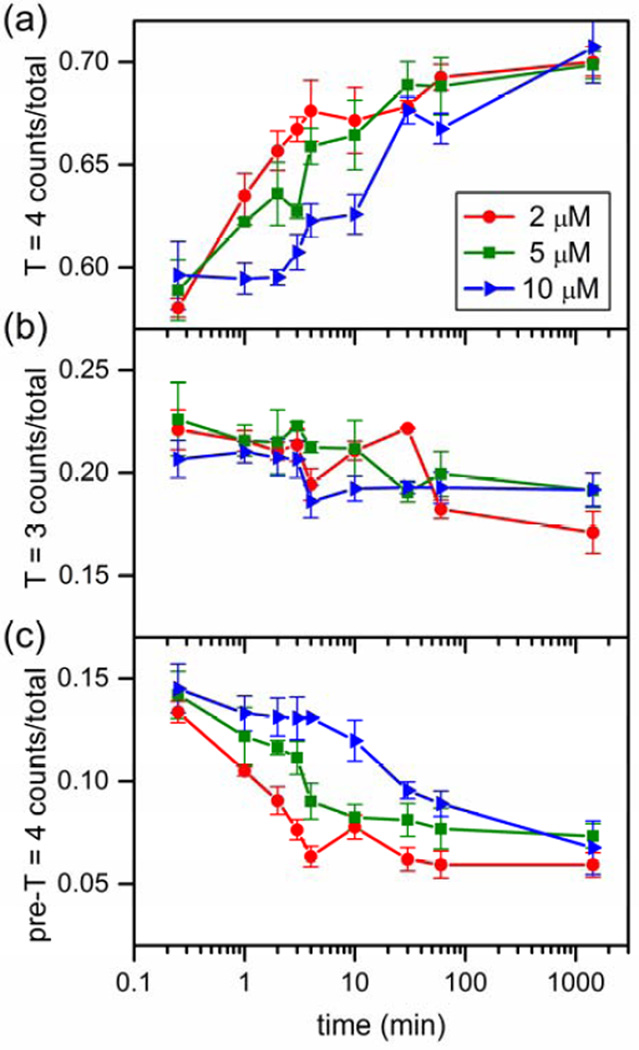
Annealing of pre-T = 4 intermediates into intact capsids over time was observed for 2, 5, and 10 µM dimer (Figure 9). At 2 µM dimer, the amount of T = 4 capsid increases as the distribution of intermediate particles is depleted. The slight decrease in the counts for T = 3 capsids could be due to completion of intermediates that are similar in size to T = 3 capsids. The growth of T = 4 capsids corresponds to depletion of late-stage intermediates centered at 105-dimers, which supports the hypothesis that these intermediates chase into T = 4 capsids. The time to anneal pre- T = 4 intermediates into T = 4 capsids is inversely proportional to dimer concentration. Growth of T = 4 capsids is continuous for assembly with 2 µM dimer, whereas 30 min are needed for the intermediates to anneal for reactions with 10 µM dimer. Slow annealing is consistent with a rate limiting step of disassembly of kinetically trapped oligomers.
Figure 9.
Variation of the fraction of (a) T = 4 capsids, (b) T = 3 capsids, and (c) pre-T = 4 intermediates with time for initial Cp149 dimer concentrations of 2, 5, and 10 µM assembled in 1 M NaCl. (a) The fraction of T = 4 capsid increases with time, and there is a corresponding decrease in the pre-T = 4 intermediate with time (panel (c)). (b) The fraction of T = 3 capsid decreases slightly over 24 h. Each trajectory is an average of at least three independent assembly reactions, and error bars are ± σ/2. Total counts are > 80,000, 240,000, and 154,000 particles for 2, 5, and 10 µM dimer, respectively.
Discussion
In these single-particle studies, we observe intermediates that transiently accumulate; these intermediates are not detected by methods such as light scattering and SEC. Theoretical models of assembly reactions predict relatively high concentrations of intermediates early in the reaction that are depleted as the assembly reaction progresses,16,18–21,40 which is in agreement with our findings. At dimer concentrations near the pseudo-critical concentration (0.5 – 1 µM), we observed robust capsid formation with a transient, low concentration of intermediates (Figure S3 in the Supporting Information). At concentrations below the pseudo-critical concentration (0.05 – 0.3 µM), assembly also occurred, but with an array of smaller intermediates (Figure 6). The presence of assembly at very low dimer concentration (50 nM) is consistent with the non-linear nature of the concentration dependence of 90- and 120-subunit capsid assembly. For a T = 4 HBV capsid, the equilibrium expression is: Kcapsid = [capsid]/[dimer]120 with the equilibrium constant having the unusual units of M−119. Because of the large exponent, virus assembly is predicted to have a pseudo-critical concentration.40 Unlike a true critical concentration, some polymerization is anticipated below the pseudo-critical concentration, and the subunit (i.e., HBV Cp149 dimer) concentration is not perfectly constant above the pseudo-critical concentration.
At relatively high dimer concentrations (≥ 2 µM dimer), capsid assembly was very rapid, and large oligomers were kinetically trapped on their way to forming T = 4 capsids (Figures 8 and 9). The window for assembly without kinetic traps is a complex function of the concentration, association energy, and the ratio of nucleation and elongation rate;18,21,41 for assembly in 1 M NaCl, 1 µM dimer is within that window, and 2 µM dimer is not. At high initial dimer concentrations, accumulation of late intermediates correlates with stalling of the assembly reaction that is progressively more apparent at higher protein concentrations. At the highest dimer concentration tested (10 µM), the reaction appears to stall for almost 30 min until intermediates anneal, rearrange, or ripen to form T = 4 capsids (Figure 9). These intermediates are consistent with late-stage assembly intermediates previously identified under kinetic trapping conditions.30 Thus, we observe that assembly at relatively high association energy is far less robust with respect to dimer concentration than at low association energy.
The sensitivity of resistive-pulse sensing facilitates a new view of the switch between T = 3 and T = 4 capsids. We observed that the ratio of T = 3 to T = 4 capsids increased with increasing the concentration of NaCl (Figure 3). One explanation for the adjustable ratio of T = 3 to T = 4 capsids is that ionic strength modifies subunit dynamic properties, inter-subunit geometry, or both.42–44 Modifications to the HBV dimer, including mutations, truncations of the C-terminus, and formation of a disulfide bond across the intra-dimer interface all change the ratio of T = 3 to T = 4 capsids.10,38 However, if structural features of the dimer are the whole story, then T = 3 and T = 4 capsids should form continuously with parallel assembly kinetics.
However, assembly kinetics for T = 3 and T = 4 capsids are completely different. Across all assembly conditions tested, T = 3 capsid assembly was complete by the time the measurement was initiated (15 s at dimer concentrations ≥ 2 µM and 90 s at dimer concentrations < 2 µM), whereas T = 4 capsids continued to assemble for minutes to hours. The critical points are that T = 3 capsids formed early in the reaction, and no appreciable T = 3 capsid formed subsequent to the initial observation.
A second hypothesis for the switch between T = 3 and T = 4 capsid assembly lies in the evolution of the energy surface during a reaction. Simulations of assembly reactions in which all intermediates are considered indicate that the selection of intermediates changes over time from those intermediates that are most kinetically accessible to intermediates that are more stable and persistent.45–46 Based on their stability and probability of formation, only a small subset of intermediates are likely to be very common.45–46 These intermediates are the steady-state backbone of virus capsid assembly kinetics.18–19,21,45,47 We speculate that among the kinetically accessible, temporally early intermediates in HBV assembly, there are one or more intermediates that provide a path to T = 3 capsids. This kinetic phenomenon might explain why a much higher T = 3 population is observed at very low dimer concentrations, well below the pseudo-critical concentration (Figures 4b and 6). At longer times, the concentrations of these T = 3 capsid predecessors collapse along with other relatively unstable intermediates, and only paths to T = 4 capsids are accessible. This proposal is consistent with theory of assembly of spherical particles, and in fact, data presented in this paper show intermediates observed at later times lead almost exclusively to T = 4 capsids (Figures 7 and 9).
Conclusion
Resistive-pulse sensing with two pores in series has excellent signal-to-noise ratios and limits of detection. With this technique, we were able to investigate the real-time assembly of HBV dimer at concentrations and with detail not previously possible. We observed assembly of Cp149 dimer below the pseudo-critical dimer concentration, revealing the feature of the T = 3 and T = 4 switch as well as the transient presence of temporally early intermediates. Equal amounts of T = 3 and T = 4 capsids are formed at the lowest concentration that assembly could be initiated, 50 nM dimer. Above the pseudo-critical dimer concentration and at high dimer concentrations, kinetic traps form that are consistent with structurally and temporally late intermediates.
Methods
Virus Capsids
HBV capsids were assembled from core protein (Cp149, 17 kDa) dimers expressed in E. coli and purified as described previously.48 Preparation of the Cp149 dimer is described in the Supporting Information. For the assembly experiments, the dimer was used without further purification. To calibrate the resistive-pulse measurements, T = 3 and T = 4 capsids were purified after assembly on a 10% – 40% (w/v) continuous sucrose gradient in 50 mM HEPES (pH 7.5) with 0.3 M NaCl that was centrifuged for 6 h at 150,000 g. The upper particle band (T = 3 capsids) and the lower particle band (T = 4 capsids) were extracted and dialyzed into 50 mM HEPES (pH 7.5) with 1 M NaCl and concentrated to 0.2 – 0.3 mg/mL. To determine sample purity for the calibration standards and capsid morphology for the products from assembly reactions, samples were adsorbed to glow-discharged carbon-coated grids (EM Sciences), stained with 2% uranyl acetate, and analyzed by transmission electron microscopy (TEM, JEM-1010, JEOL Ltd.).
Device Fabrication
The nanofluidic devices were fabricated in two steps. First, the microchannels were fabricated in D263 glass substrates by standard UV photolithography and wet chemical etching.34 After microchannel fabrication, the nanochannel and nanopores were milled directly into the glass substrates with a focused ion beam (FIB) instrument (Auriga 60, Carl Zeiss, Inc.) controlled by NanoPatterning and Visualization Engine (FIBICS, Inc.). The nanochannel sections that connected the pores with the microchannels were milled with a 30-kV beam at 50 pA and a dose of 1 nC/µm2. The pore-to-pore channel was milled with the same accelerating potential and beam current, but with a dose of 0.5 nC/µm2. Last, the two nanopores were milled as a single line pass with a 30-kV beam at 20 pA and a dose of 0.006 µC/µm to connect the three nanochannel sections. During the FIB milling, an electron flood gun (FG 15/40, SPECS, GmbH) was operated at 5 eV and 20 µA to compensate for the build-up of positive charge on the substrate surface. The nanochannel and nanopore dimensions were determined with the scanning electron microscope (SEM) on the FIB instrument and an atomic force microscope (AFM; MFP-3D, Asylum Research, Inc.). The nanochannel is composed of three sections: one 300-nm wide, 110-nm deep, and 1-µm long pore-to-pore channel that connects the two nanopores and two 500-nm wide, 220-nm deep, and 4-µm long channels that connect the nanopores to the microchannels. The nanopores are 200 ± 10 nm long, 60 ± 5 nm wide, and 60 ± 5 nm deep (Figure 1b).
To bond the devices, the substrates and #1.5 cover glass were hydrolyzed in a solution of 1 M NaOH for 15 min at room temperature, sonicated in water, brought into contact with each other, dried overnight at 90 °C, and annealed in a furnace at 545 °C for 10 h. After bonding, a 1 M NaOH solution was flushed through the nanochannel for 60 to 90 min to etch the nanopores to a final width and depth of 60 ± 5 nm to allow passage of large aggregates formed during the assembly reactions. The assembly experiments were performed on six devices.
Resistive-Pulse Measurements
Four types of resistive-pulse measurements are reported: (1) calibration of the nanopores, (2) assembly at low initial dimer concentration (< 2 µM), (3) assembly at high initial dimer concentration (≥ 2 µM), and (4) assembly at different NaCl concentrations. Prior to each assembly experiment, we calibrated each device with purified T = 3 and T = 4 capsids in 50 mM HEPES buffer (pH 7.5) with 1 M NaCl (Figure S1 in the Supporting Information). For assembly reactions at low dimer concentrations (< 2 µM), Cp149 dimer was brought to the designated initial dimer concentration in 50 mM HEPES buffer (pH 7.5) with 1 M NaCl and loaded onto the device. A period of 90 s elapsed between initial mixing of the dimer into 1 M NaCl to initiate assembly and the start of the resistive-pulse measurement. For assembly reactions at high dimer concentrations (≥ 2 µM), Cp149 dimer was brought to the designated initial dimer concentration in 50 mM HEPES buffer with 1 M NaCl to initiate assembly, the assembly reaction was allowed to proceed for the designated time, and the reaction mixture was diluted to an equivalent 0.25 or 0.5 µM dimer in 50 mM HEPES buffer with 1 M NaCl prior to being loaded onto the device for resistive-pulse measurements. For assembly at different NaCl concentrations, Cp149 dimer was brought to the designated initial dimer concentration in 50 mM HEPES buffer (pH 7.5) with 0.3, 0.5, 0.7, or 1 M NaCl, and the assembly reaction proceeded for 90 min. At 90 min, the reaction mixture was diluted to an equivalent 0.5 µM dimer in 50 mM HEPES buffer with 1 M NaCl prior to being loaded onto the device for resistive-pulse measurements.
After samples were loaded into the sample reservoir, an Axopatch 200B current amplifier (Molecular Devices, Inc.) was used to apply a potential (450 to 550 mV) between the sample and waste reservoirs (Figure 1a) to drive the sample electrokinetically through the nanochannel. The amplifier also measured the current through the nanochannel at a filter frequency of 10 kHz and a collection frequency of 40 kHz. The potentials were applied with Ag/AgCl electrodes placed in the buffer-filled reservoirs, and the electrical measurements were conducted inside a Faraday cage covered in acoustic wedge foam.
Current data were imported into MatLab R2014a (Mathworks, Inc.), and a modified version of Open Nanopore 1.2 was used to fit the raw data to determine the pulse amplitude (Δi), pulse width (w), and average baseline current adjacent to each pulse.49 Times between pulses were tabulated and plotted on a log scale to reveal two distributions of the pore-to-pore times for individual capsids and uncorrelated times between pulses from different capsids. A Gaussian function was fitted to the pore-to-pore time (tpp) distribution, and ±2σ from the fit was used as a selection criterion for related pulse pairs. The average pulse amplitude from each pulse pair was divided by average baseline current adjacent to each pulse to calculate Δi/i, which was then plotted as a histogram with a bin size of 1 × 10−4. Gaussian functions were fitted to the Δi/i distributions to determine the T = 3 capsid, T = 4 capsid, pre-T = 4 intermediate, and non-capsid oligomer distributions.
Supplementary Material
Acknowledgments
This work was supported in part by NIH R01 GM100071 and NSF CHE-0923064 for Z.D.H and S.C.J. and by NIH R01 GM100071 and NIH R56 AI077688 for L.S. and A.Z. The authors thank the Indiana University Nanoscale Characterization Facility for use of its instruments.
Footnotes
Supporting Information Available. Sections for materials, preparation and purification of Cp149 dimer, calibration of nanofluidic devices, calibration of capsid concentration, assembly near the pseudo-critical dimer concentration, transmission electron microscopy, and elongation rate. This material is available free of charge via the Internet at http://pubs.acs.org.
A.Z. reports a financial interest in a company based on assembly effectors.
References
- 1.Zlotnick A, Mukhopadhyay S. Virus Assembly, Allostery and Antivirals. Trends Microbiol. 2011;19:14–23. doi: 10.1016/j.tim.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Prevelige PE. Inhibiting Virus-Capsid Assembly by Altering the Polymerisation Pathway. Trends Biotech. 1998;16:61–65. doi: 10.1016/s0167-7799(97)01154-2. [DOI] [PubMed] [Google Scholar]
- 3.Zlotnick A, Stray SJ. How Does Your Virus Grow? Understanding and Interfering with Virus Assembly. Trends Biotech. 2003;21:536–542. doi: 10.1016/j.tibtech.2003.09.012. [DOI] [PubMed] [Google Scholar]
- 4.Klumpp K, Crepin T. Capsid Proteins of Enveloped Viruses as Antiviral Drug Targets. Curr. Opin. Virol. 2014;5:63–71. doi: 10.1016/j.coviro.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 5.Fischlechner M, Donath E. Viruses as Building Blocks for Materials and Devices. Angew. Chem. Int. Ed. 2007;46:3184–3193. doi: 10.1002/anie.200603445. [DOI] [PubMed] [Google Scholar]
- 6.Douglas T, Young M. Viruses: Making Friends with Old Foes. Science. 2006;312:873–875. doi: 10.1126/science.1123223. [DOI] [PubMed] [Google Scholar]
- 7.Zlotnick A, Venkatakrishnan B, Tan Z, Lewellyn EB, Turner WW, Francis S. Core Protein: A Pleiotropic Keystone in the Hbv Lifecycle. Antiviral Res. 2015 doi: 10.1016/j.antiviral.2015.06.020. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Papatheodoridis G, Buti M, Cornberg M, Janssen H, Mutimer D, Pol S, Raimondo G. Easl Clinical Practice Guidelines: Management of Chronic Hepatitis B Virus Infection. J. Hepatol. 2012;57:167–185. doi: 10.1016/j.jhep.2012.02.010. [DOI] [PubMed] [Google Scholar]
- 9.Crowther RA, Kiselev NA, Bottcher B, Berriman JA, Borisova GP, Ose V, Pumpens P. 3-Dimensional Structure of Hepatitis-B Virus Core Particles Determined by Electron Cryomicroscopy. Cell. 1994;77:943–950. doi: 10.1016/0092-8674(94)90142-2. [DOI] [PubMed] [Google Scholar]
- 10.Zlotnick A, Cheng N, Conway JF, Booy FP, Steven AC, Stahl SJ, Wingfield PT. Dimorphism of Hepatitis B Virus Capsids Is Strongly Influenced by the C-Terminus of the Capsid Protein. Biochemistry. 1996;35:7412–7421. doi: 10.1021/bi9604800. [DOI] [PubMed] [Google Scholar]
- 11.Ceres P, Zlotnick A. Weak Protein-Protein Interactions Are Sufficient to Drive Assembly of Hepatitis B Virus Capsids. Biochemistry. 2002;41:11525–11531. doi: 10.1021/bi0261645. [DOI] [PubMed] [Google Scholar]
- 12.Kenney JM, Vonbonsdorff CH, Nassal M, Fuller SD. Evolutionary Conservation in the Hepatitis-B Virus Core Structure - Comparison of Human and Duck Cores. Structure. 1995;3:1009–1019. doi: 10.1016/s0969-2126(01)00237-4. [DOI] [PubMed] [Google Scholar]
- 13.Dryden KA, Wieland SF, Whitten-Bauer C, Gerin JL, Chisari FV, Yeager M. Native Hepatitis B Virions and Capsids Visualized by Electron Cryomicroscopy. Mol. Cell. 2006;22:843–850. doi: 10.1016/j.molcel.2006.04.025. [DOI] [PubMed] [Google Scholar]
- 14.Seitz S, Urban S, Antoni C, Boettcher B. Cryo-Electron Microscopy of Hepatitis B Virions Reveals Variability in Envelope Capsid Interactions. EMBO J. 2007;26:4160–4167. doi: 10.1038/sj.emboj.7601841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zlotnick A, Johnson JM, Wingfield PW, Stahl SJ, Endres D. A Theoretical Model Successfully Identifies Features of Hepatitis B Virus Capsid Assembly. Biochemistry. 1999;38:14644–14652. doi: 10.1021/bi991611a. [DOI] [PubMed] [Google Scholar]
- 16.Hagan MF, Elrad OM. Understanding the Concentration Dependence of Viral Capsid Assembly Kinetics-the Origin of the Lag Time and Identifying the Critical Nucleus Size. Biophys. J. 2010;98:1065–1074. doi: 10.1016/j.bpj.2009.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zandi R, van der Schoot P, Reguera D, Kegel W, Reiss H. Classical Nucleation Theory of Virus Capsids. Biophys. J. 2006;90:1939–1948. doi: 10.1529/biophysj.105.072975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morozov AY, Bruinsma RF, Rudnick J. Assembly of Viruses and the Pseudo-Law of Mass Action. J. Chem. Phys. 2009;131:155101. doi: 10.1063/1.3212694. [DOI] [PubMed] [Google Scholar]
- 19.Katen S, Zlotnick A. Thermodynamics of Virus Capsid Assembly. Methods Enzymol. 2009;455:395–417. doi: 10.1016/S0076-6879(08)04214-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zlotnick A. Theoretical Aspects of Virus Capsid Assembly. J. Mol. Recognit. 2005;18:479–490. doi: 10.1002/jmr.754. [DOI] [PubMed] [Google Scholar]
- 21.Endres D, Zlotnick A. Model-Based Analysis of Assembly Kinetics for Virus Capsids or Other Spherical Polymers. Biophys. J. 2002;83:1217–1230. doi: 10.1016/S0006-3495(02)75245-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Prevelige PE, Thomas D, King J. Nucleation and Growth Phases in the Polymerization of Coat and Scaffolding Subunits into Icosahedral Procapsid Shells. Biophys. J. 1993;64:824–835. doi: 10.1016/S0006-3495(93)81443-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zlotnick A, Aldrich R, Johnson JM, Ceres P, Young MJ. Mechanism of Capsid Assembly for an Icosahedral Plant Virus. Virology. 2000;277:450–456. doi: 10.1006/viro.2000.0619. [DOI] [PubMed] [Google Scholar]
- 24.Johnson JM, Tang JH, Nyame Y, Willits D, Young MJ, Zlotnick A. Regulating Self-Assembly of Spherical Oligomers. Nano Lett. 2005;5:765–770. doi: 10.1021/nl050274q. [DOI] [PubMed] [Google Scholar]
- 25.Chen C, Kao CC, Dragnea B. Self-Assembly of Brome Mosaic Virus Capsids: Insights from Shorter Time-Scale Experiments. J. Phys. Chem. A. 2008;112:9405–9412. doi: 10.1021/jp802498z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Casini GL, Graham D, Heine D, Garcea RL, Wu DT. In vitro Papillomavirus Capsid Assembly Analyzed by Light Scattering. Virology. 2004;325:320–327. doi: 10.1016/j.virol.2004.04.034. [DOI] [PubMed] [Google Scholar]
- 27.Kler S, Asor R, Li CL, Ginsburg A, Harries D, Oppenheim A, Zlotnick A, Raviv U. Rna Encapsidation by Sv40-Derived Nanoparticles Follows a Rapid Two-State Mechanism. J. Am. Chem. Soc. 2012;134:8823–8830. doi: 10.1021/ja2110703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tresset G, Le Coeur C, Bryche J-F, Tatou M, Zeghal M, Charpilienne A, Poncet D, Constantin D, Bressanelli S. Norovirus Capsid Proteins Self-Assemble through Biphasic Kinetics Via Long-Lived Stave-Like Intermediates. J. Am. Chem. Soc. 2013;135:15373–15381. doi: 10.1021/ja403550f. [DOI] [PubMed] [Google Scholar]
- 29.Uetrecht C, Barbu IM, Shoemaker GK, van Duijn E, Heck AJR. Interrogating Viral Capsid Assembly with Ion Mobility-Mass Spectrometry. Nat. Chem. 2011;3:126–132. doi: 10.1038/nchem.947. [DOI] [PubMed] [Google Scholar]
- 30.Pierson EE, Keifer DZ, Selzer L, Lee LS, Contino NC, Wang JCY, Zlotnick A, Jarrold MF. Detection of Late Intermediates in Virus Capsid Assembly by Charge Detection Mass Spectrometry. J. Am. Chem. Soc. 2014;136:3536–3541. doi: 10.1021/ja411460w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Borodavka A, Tuma R, Stockley PG. Evidence That Viral Rnas Have Evolved for Efficient, Two-Stage Packaging. Proc. Natl. Acad. Sci. U. S. A. 2012;109:15769–15774. doi: 10.1073/pnas.1204357109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou K, Li L, Tan Z, Zlotnick A, Jacobson SC. Characterization of Hepatitis B Virus Capsids by Resistive-Pulse Sensing. J. Am. Chem. Soc. 2011;133:1618–1621. doi: 10.1021/ja108228x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harms ZD, Mogensen KB, Nunes PS, Zhou K, Hildenbrand BW, Mitra I, Tan Z, Zlotnick A, Kutter JP, Jacobson SC. Nanofluidic Devices with Two Pores in Series for Resistive-Pulse Sensing of Single Virus Capsids. Anal. Chem. 2011;83:9573–9578. doi: 10.1021/ac202358t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harms ZD, Haywood DG, Kneller AR, Selzer L, Zlotnick A, Jacobson SC. Single-Particle Electrophoresis in Nanochannels. Anal. Chem. 2015;87:699–705. doi: 10.1021/ac503527d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Uram JD, Ke K, Hunt AJ, Mayer M. Label-Free Affinity Assays by Rapid Detection of Immune Complexes in Submicrometer Pores. Angew. Chem., Int. Ed. 2006;45:2281–2285. doi: 10.1002/anie.200502862. [DOI] [PubMed] [Google Scholar]
- 36.Balakrishnan KR, Anwar G, Chapman MR, Nguyen T, Kesavaraju A, Sohn LL. Node-Pore Sensing: A Robust, High-Dynamic Range Method for Detecting Biological Species. Lab Chip. 2013;13:1302–1307. doi: 10.1039/c3lc41286e. [DOI] [PubMed] [Google Scholar]
- 37.McMullen A, de Haan HW, Tang JX, Stein D. Stiff Filamentous Virus Translocations through Solid-State Nanopores. Nat. Comm. 2014;5 doi: 10.1038/ncomms5171. [DOI] [PubMed] [Google Scholar]
- 38.Selzer L, Katen SP, Zlotnick A. The Hepatitis B Virus Core Protein Intradimer Interface Modulates Capsid Assembly and Stability. Biochemistry. 2014;53:5496–5504. doi: 10.1021/bi500732b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zlotnick A, Tan Z, Selzer L. One Protein, at Least Three Structures, and Many Functions. Structure. 2013;21:6–8. doi: 10.1016/j.str.2012.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zlotnick A. To Build a Virus Capsid - an Equilibrium Model of the Self-Assembly of Polyhedral Protein Complexes. J. Mol. Biol. 1994;241:59–67. doi: 10.1006/jmbi.1994.1473. [DOI] [PubMed] [Google Scholar]
- 41.Hagan MF, Elrad OM, Jack RL. Mechanisms of Kinetic Trapping in Self- Assembly and Phase Transformation. J. Chem. Phys. 2011;135:104115. doi: 10.1063/1.3635775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Siber A, Majdandzic A. Spontaneous Curvature as a Regulator of the Size of Virus Capsids. Phys. Rev. E. 2009;80 doi: 10.1103/PhysRevE.80.021910. [DOI] [PubMed] [Google Scholar]
- 43.Elrad OM, Hagan MF. Mechanisms of Size Control and Polymorphism in Viral Capsid Assembly. Nano Lett. 2008;8:3850–3857. doi: 10.1021/nl802269a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reddy VS, Giesing HA, Morton RT, Kumar A, Post CB, Brooks CL, Johnson JE. Energetics of Quasiequivalence: Computational Analysis of Protein- Protein Interactions in Icosahedral Viruses. Biophys. J. 1998;74:546–558. doi: 10.1016/S0006-3495(98)77813-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Endres D, Miyahara M, Moisant P, Zlotnick A. A Reaction Landscape Identifies the Intermediates Critical for Self-Assembly of Virus Capsids and Other Polyhedral Structures. Protein Sci. 2005;14:1518–1525. doi: 10.1110/ps.041314405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moisant P, Neeman H, Zlotnick A. Exploring the Paths of (Virus) Assembly. Biophys. J. 2010;99:1350–1357. doi: 10.1016/j.bpj.2010.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Keef T, Micheletti C, Twarock R. Master Equation Approach to the Assembly of Viral Capsids. J. Theor. Biol. 2006;242:713–721. doi: 10.1016/j.jtbi.2006.04.023. [DOI] [PubMed] [Google Scholar]
- 48.Zlotnick A, Ceres P, Singh S, Johnson JM. A Small Molecule Inhibits and Misdirects Assembly of Hepatitis B Virus Capsids. J. Virol. 2002;76:4848–4854. doi: 10.1128/JVI.76.10.4848-4854.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Raillon C, Granjon P, Graf M, Steinbock LJ, Radenovic A. Fast and Automatic Processing of Multi-Level Events in Nanopore Translocation Experiments. Nanoscale. 2012;4:4916–4924. doi: 10.1039/c2nr30951c. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.