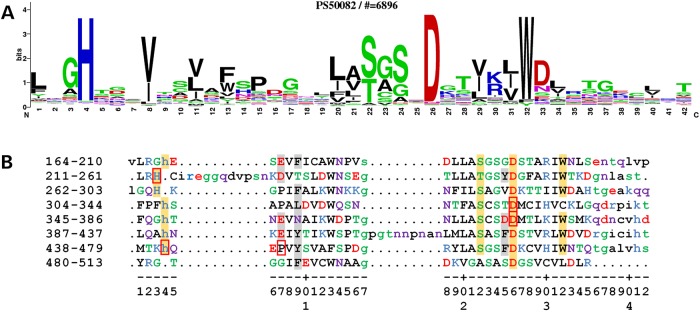
Figure 2.
Conserved sequence elements of the WD40 motif. (A) PROSITE sequence logo for the WD40 motif, derived from a multiple sequence alignment of 6896 sequence fragments. The one-letter amino acid codes are coloured by type (blue basic, red acidic, green and purple polar and the rest black). The height of each corresponds to its frequency of occurrence in the alignment. (B) Structure-based alignment of the eight WD40 motifs in the crystal structure of TBLR1. The motifs were manually extracted from the 4lg9 PDB file and then aligned using the PDBeFold Server (36). The numbers on the left show the range of residue numbers in the sequence on that line. The one-letter amino acid codes are coloured as per the PROSITE sequence logo (A); lower-case letters correspond to residues not aligned in the 3D superposition. The numbers along the bottom roughly correspond to the sequence positions in the WD40 motif in (A). The amino acids having an orange background are those belonging to the Asp-His-Ser/Thr-Trp tetrad. The red borders identify the five amino acids involved in the DDD missense mutations: His213Gln, Asp328Gly, Asp370Tyr, His441Arg and Pro444Arg. The amino acids with the light grey backgrounds are the hotspot residues on the domain's top face, as identified by WDSPdb (31), being the ones likely to interact with β-catenin when it binds.