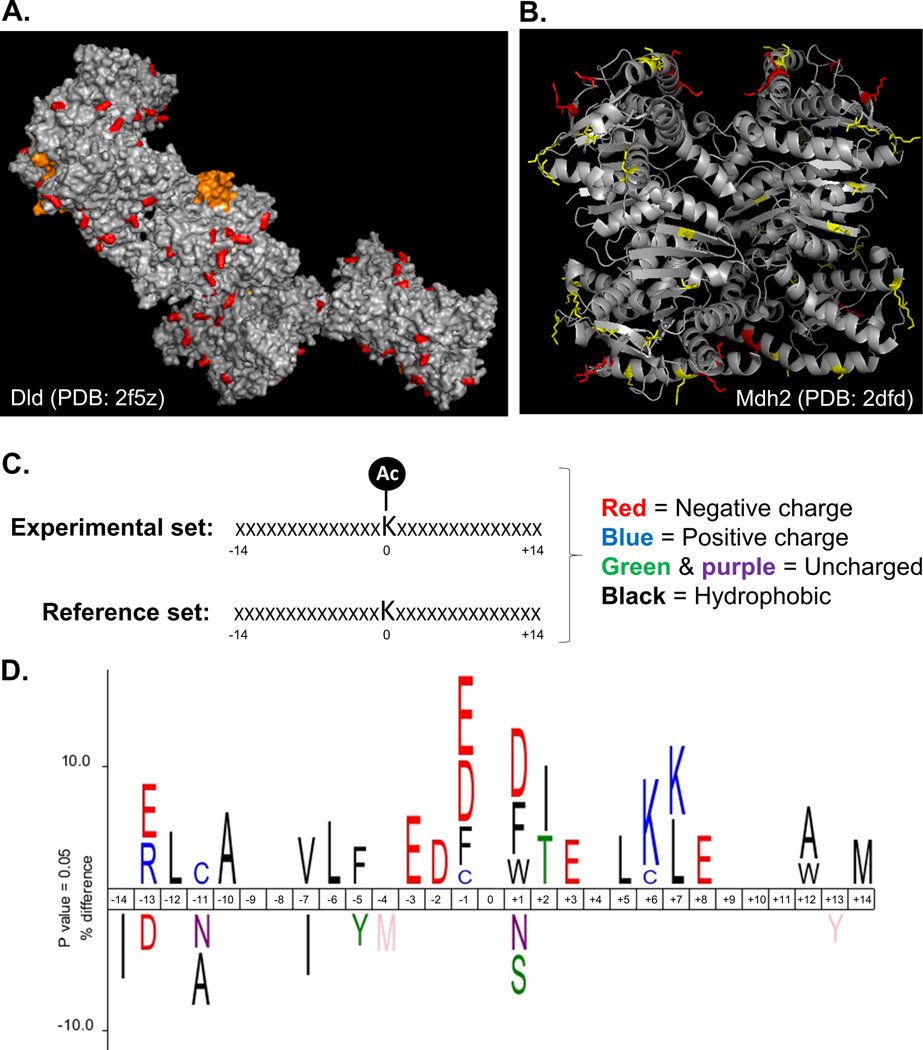
Figure 5. In silico structural analysis and modeling of acetylpeptides and proteins increased by CrAT deficiency.
(a) Structure (PDB: 2f5z) of the E3 complex of pyruvate dehydrogenase (Dld) showing acetylated lysine residues (AcK) that increased in the CrATskm−/− (highlighted in red). Residues in orange represent the FAD-linked lipoamide arm of the enzyme. Modeling was performed using PyMOL. (b) Structure (PDB: 2dfd) of mitochondrial malate dehydrogenase 2 (Mdh2) showing localization of acetylated residues. AcK highlighted in red represent differentially abundant PTMs identified from CrATskm−/− mice, whereas those in yellow were unchanged between genotypes. (c) Icelogo analysis was performed by comparing the amino acid landscape surrounding AcKs identified in CrATskm−/− muscle to a mouse reference set containing information about the identity of the amino acids in the same 28 positions neighboring lysine residues in all mouse proteins. (d) The top results show the enrichment of amino acids seven residues N- (negative) or C- (positive) terminal to acetylated lysines (centered at position “0”) identified to be increased in CrATskm−/− muscle compared to controls after 12 weeks of high fat diet feeding. The bottom results show the enrichment of amino acids surrounding the average lysine present in mouse proteins. Letters are color coded according to amino acid type using the scheme shown in panel c.