Figure 1. Genetically Engineered Mice with InsG3680 or R1117X Shank3 Mutation Differentially Express SHANK3 Protein and mRNA.
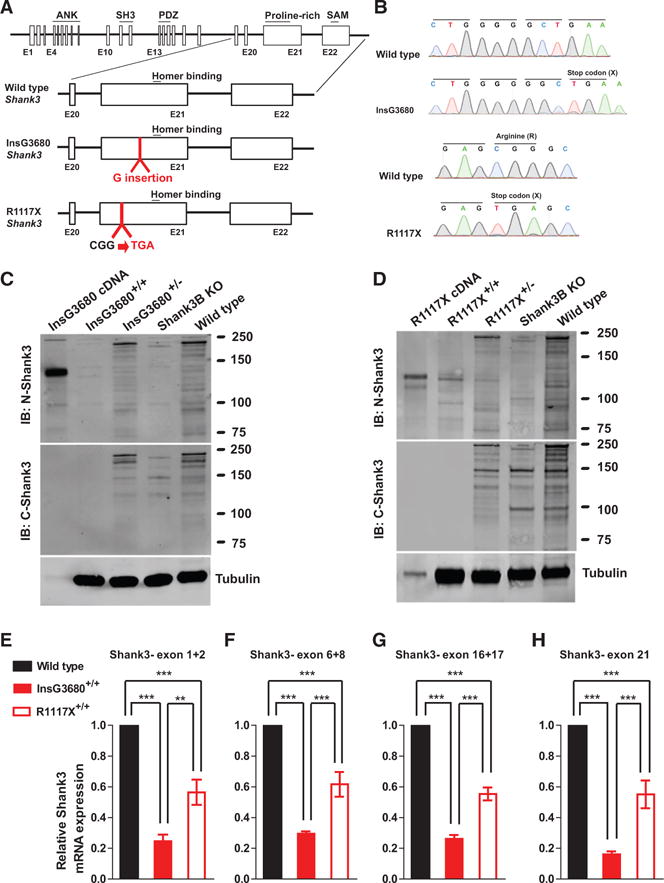
(A) Schematic diagram for wild-type Shank3-, InsG3680-, and R1117X-targeted Shank3 alleles. Gene structure of wild-type mouse Shank3 gene and magnified panels on the structure between exon 20 and exon 22 are shown below. Top: wild-type Shank3; middle: autism-associated InsG3680 Shank3 mutation with an insertion of “guanine” nucleotide at position 3680; bottom: schizophrenia-associated R1117X Shank3 mutation changing the “CGG” codon for arginine to a “TGA” stop codon.
(B) Representative sequencing chromatograms of wild-type and InsG3680 mutated alleles; wild-type and R1117X mutated alleles showing the point mutations.
(C) Representative western blots using striatal PSD fractions prepared from wild-type, Shank3B KO mice, InsG3680+/− mice, InsG3680+/+ mice; lysate from HEK239 cells expressing cDNA plasmid encoding the InsG3680 mutated Shank3. Note that neither the antibody against the N nor C terminus detected SHANK3 protein in InsG3680+/+ mice.
(D) Representative western blots using striatal PSD fractions prepared from wild-type, Shank3B KO mice, R1117X+/− mice, R1117X+/+ mice; lysate from HEK239 cells expressing cDNA plasmid encoding the R1117X mutated Shank3. Note that SHANK3 protein expression is almost abolished except for a prominent truncated isoform in the R1117X+/+ line that is detected with the antibody against the N terminus.
(E–H) The relative level of Shank3 mRNA in striatum is different between the two lines as quantified by primers amplifying exon 1 to 2, exon 6 to 8, 16 to 17, and partial exon 21 before both Shank3 mutation sites. Data are normalized to Gapdh mRNA and presented as mean ± SEM. **p < 0.01, ***p < 0.001; one-way ANOVA with Bonferroni post hoc test, WT mice (n = 5), R1117X+/+ mice (n = 5) and InsG3680+/+ mice (n = 5).
