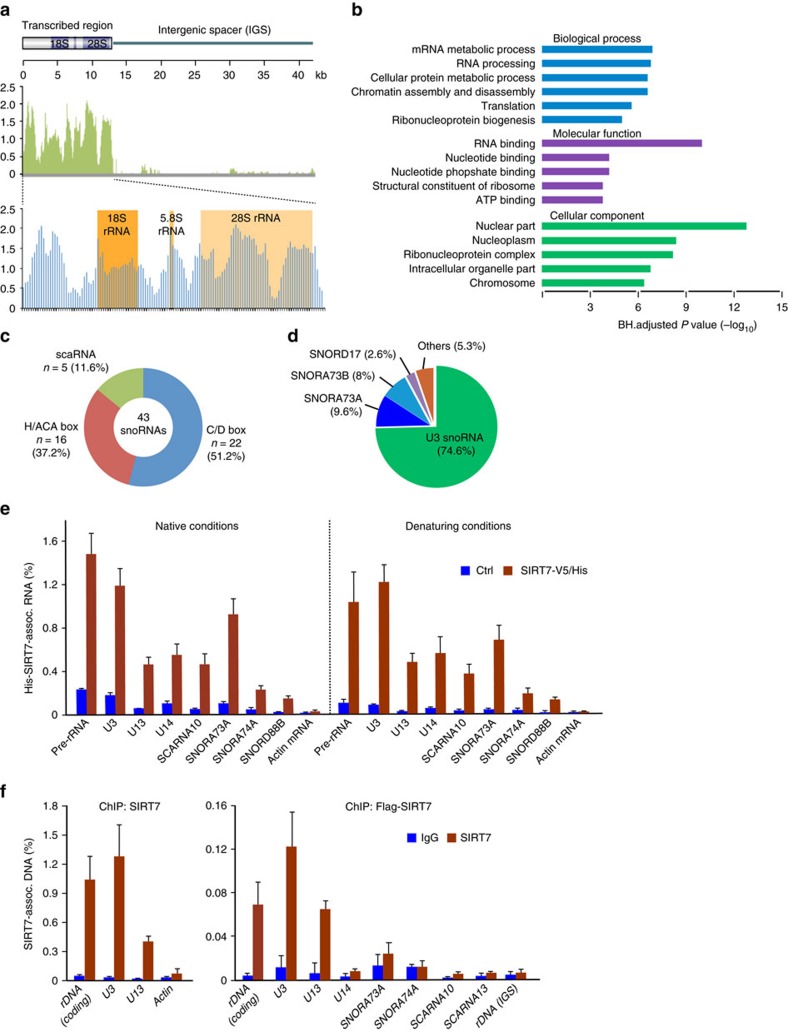
Figure 1. SIRT7 is associated with pre-rRNA and snoRNAs.
(a) SIRT7 CLIP-seq reads mapped to a custom annotation file of a human rDNA repeat (middle) or the transcribed region (bottom). The region encoding 18S, 5.8S and 28S rRNA is highlighted. SIRT7 reads after subtraction of IgG reads were normalized to input reads (y axis). (b) Gene ontology categories of SIRT7 CLIP-seq peaks. The most representative clusters are shown according to the ajusted P value (−log10). (c) SIRT7-bound snoRNAs comprise C/D box, H/ACA box snoRNAs and scaRNAs. The number (n) and relative abundance (%) of each snoRNA class associated with SIRT7 is presented. (d) U3, SNORA73A and 73B snoRNAs are overrepresented among SIRT7-associated snoRNAs. SIRT7 reads mapped to corresponding snoRNAs are indicated as percentage of all snoRNAs identified by CLIP-seq. (e) Comparison of SIRT7-associated RNAs under native and denaturing conditions. His/V5-tagged SIRT7 expressed in HEK293T cells was affinity-purified on Ni-NTA-agarose under native or denaturing conditions, and associated RNAs were detected by RT–qPCR. Lysates from non-transfected HEK293T cells were used for control (Ctrl). Associated pre-RNA was monitored by RT–qPCR using primer H1 (Supplementary Table 3). Bars represent means±s.d. from three experiments. See also Supplementary Fig. 2b,d. (f) ChIP assays showing association of endogenous SIRT7 (left panel) or transiently overexpressed Flag-SIRT7 (right panel) with the indicated gene loci in HEK293T cells. rDNA was amplified using primers H4 (coding) and H18 (IGS; Supplementary Table 3). Bars represent means±s.d. from three experiments. See also Supplementary Fig. 2d.