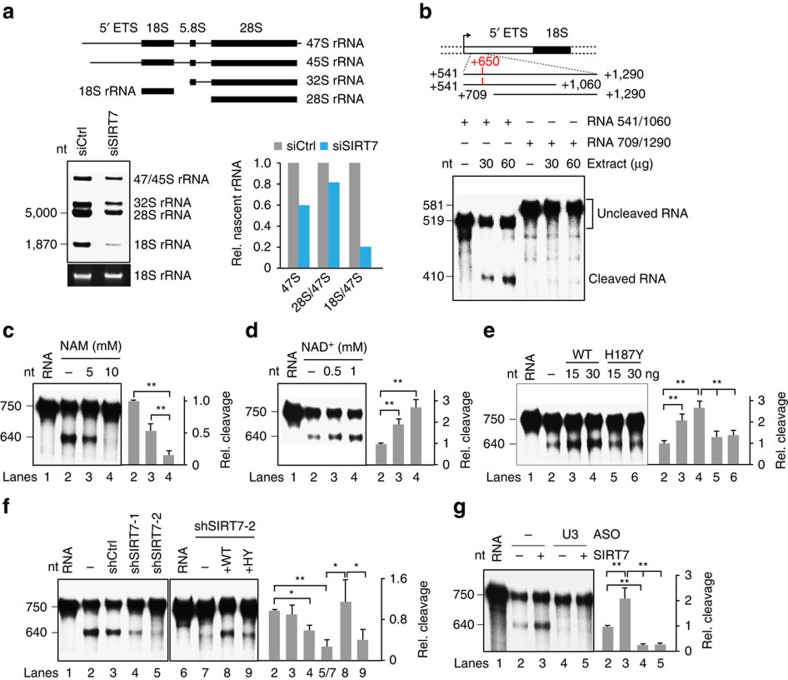
Figure 2. SIRT7 is involved in pre-rRNA processing.
(a) Knockdown of SIRT7 impairs pre-rRNA synthesis and processing in vivo. U2OS cells transfected with control (siCtrl) or SIRT7-specific siRNAs (siSIRT7) were metabolically labelled with 3H-uridine. RNA was analysed by agarose gel electrophoresis and fluorography. The bar diagram shows quantification of the processing intermediates, values from siCtrl cells being set to 1. (b) In vitro processing assay. Extracts from L1210 cells were incubated with 32P-labelled RNA comprising the 5′ETS depicted in the scheme above. 32P-labelled RNA and cleavage products were analysed by gel electrophoresis and PhosphorImaging. See also Supplementary Fig. 3a. (c) 5′ETS processing is inhibited by NAM. The assay contained radiolabelled RNA (+541/+1290) and extracts from L1210 cells cultured for 6 h in the absence or presence of NAM. (d) Processing is enhanced by NAD+. Processing assays containing radiolabelled RNA (+541/+1290) were substituted with NAD+ as indicated. (e) The catalytic activity of SIRT7 is required for pre-rRNA cleavage. Assays were supplemented with 15 or 30 ng of purified wildtype (WT) or mutant (H187Y) Flag-SIRT7 (Supplementary Fig. 3b). (f) Depletion of SIRT7 impairs processing. SIRT7 was depleted from L1210 cells by shRNAs (shSIRT7-1, shSIRT7-2, Supplementary Fig. 3c). Extracts from non-infected cells (−) or cells expressing control shRNA (shCtrl) served as control (left). To rescue impaired cleavage, 15 ng of wild-type Flag-SIRT7 (WT) or mutant H187Y (HY) were added to SIRT7-depleted extracts (right). (g) Depletion of U3 snoRNA abolishes processing. U3 snoRNA was depleted by preincubating extracts with U3-specific antisense oligos (ASO, 50 ng μl−1) and 2 U of RNase H (Supplementary Fig. 3d). In vitro processing was performed with undepleted (−) or depleted extracts in the absence or presence of 15 ng Flag-SIRT7. Bar diagrams in c–g show quantification of the ratio of cleaved versus uncleaved transcripts, presented as mean±s.d. from three independent experiments (*P<0.05, **P<0.01, analysis of variance with Bonferroni's test).