Figure 1. Conformational differences between the active protomer (protomer A) of SARS-3CLpro and the R298A mutant (drawn by PYMOL1.4.1).
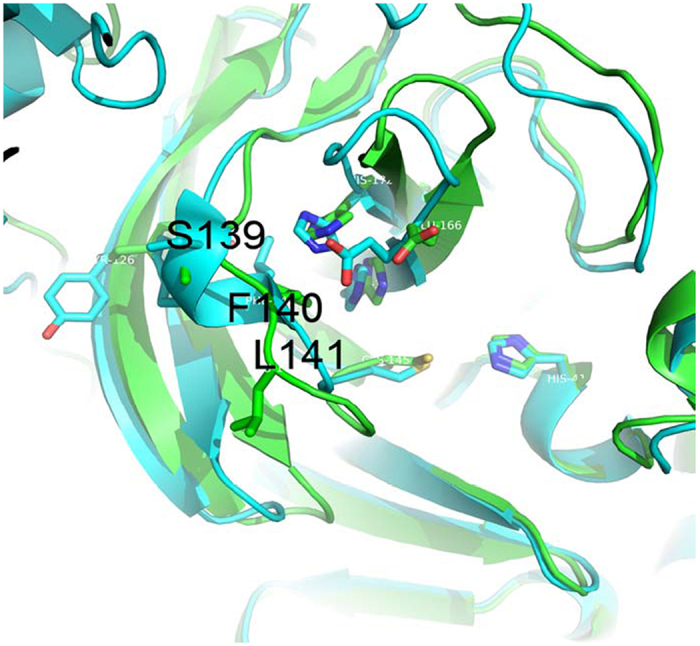
The active protomer of SARS-3CLpro (PDB ID 1UK2) and the R298A mutant (PDB ID 2QCY) are shown in green and cyan, respectively. The most distinguishable characteristic of the R298A mutant is the formation of a short 310-helix instead of an active-site loop in residues Ser139-Phe140-Leu141.
