Figure 6. Fra's transcriptional activation function is required for its ability to regulate midline crossing and comm expression.
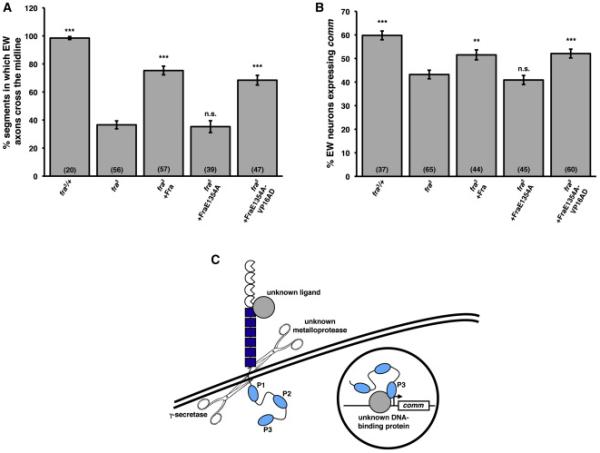
A) Quantification of EW axon crossing in stage 14 embryos. Data were analyzed by ANOVA, followed by Student's t-test. *** indicates p<0.0001, compared to fra mutants. n.s. indicates p>0.05, compared to fra mutants. Error bars indicate SEM. Number in parentheses indicates number of embryos scored. Note that the data for fra3 heterozygotes and fra3 mutants are also shown in Figures 2B and S3.
B) Quantification of comm expression in EW neurons in stage 14 embryos. Data were analyzed by ANOVA, followed by Student's t-test. *** indicates p<0.0001, compared to fra mutants. ** indicates p<0.005, compared to fra mutants. n.s. indicates p>0.05, compared to fra mutants. Error bars indicate SEM. Number in parentheses indicates number of embryos scored. Note that the data for fra3 heterozygotes and fra3 mutants are also shown in Figures 2C and S2 and that the data for fra3 mutants rescued with wild type Fra are also shown in Figure S2.
C) A model for Fra-dependent comm expression. Full-length Fra is cleaved by γ-secretase, likely in response to an unknown ligand, which stimulates metalloprotease cleavage. The soluble ICD then translocates to the nucleus, where it functions as a transcriptional activator to induce comm expression, either directly or indirectly. The Fra ICD likely associates with DNA by interacting with one or more unknown DNA-binding proteins. P3 functions as a transcriptional activation domain.
See also Figure S3.
