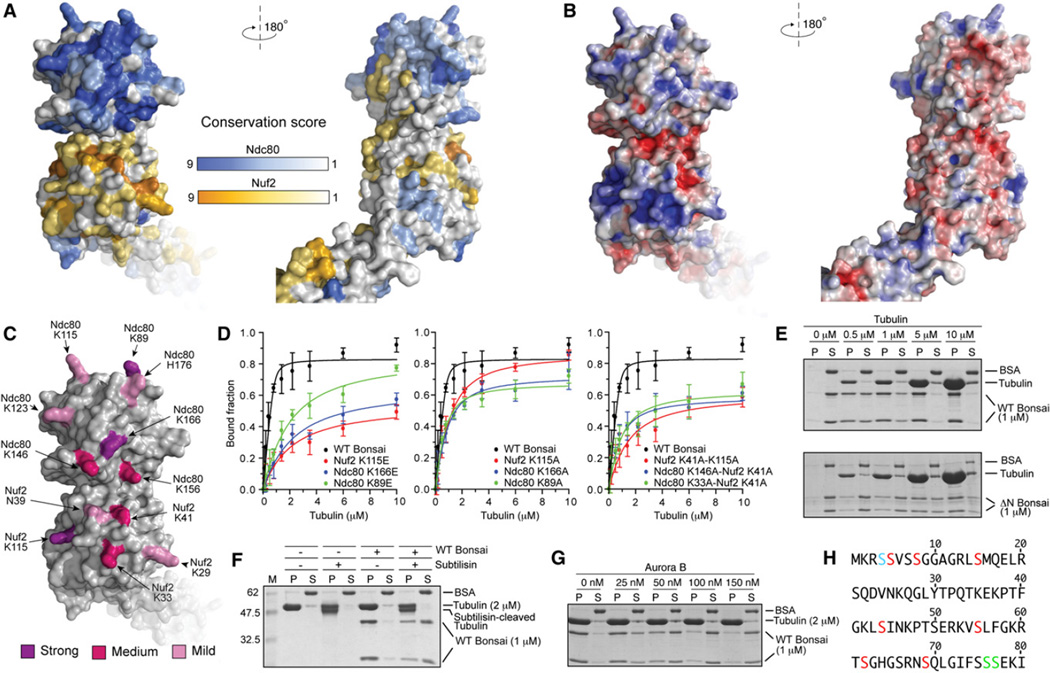
Figure 4. Organization of the Microtubule-Binding Region.
(A) Surface views of the Ndc80-Nuf2 globular region colored according to conservation.
(B) Surface views colored by electrostatic potential. The orientations are as in (A).
(C) Positions of mutated residues. Residues whose KE mutation caused a 30- to 40-fold impairment of microtubule binding are shown in purple (see also panel D, Table S1, and Figure S2). Residues whose KE mutation caused a 6- to 12-fold impairment in microtubule binding are shown in magenta. Residues whose mutation into alanine provided modest destabilization of binding that was not quantitated are shown in pink.
(D) Plot of quantifications of microtubule cosedimentation assay with Ndc80bonsai and the indicated mutants (see also Table S1 and Figure S2). Error bars represent standard deviations calculated from at least three independent experiments.
(E) The ability of Ndc80bonsai and Ndc80ΔN-bonsai to cosediment with microtubules was tested in parallel.
(F) Ndc80bonsai showed strongly reduced binding to subtilisin-treated microtubules (see Supplemental Experimental Procedures for details).
(G) Aurora B releases Ndc80bonsai from microtubules.
(H) Sequence of the N-terminal tail of Ndc80. Residues in red were phosphorylated by Aurora B in vitro (see Supplemental Experimental Procedures). Residues in green are phosphorylated in vivo but do not conform to the Aurora B consensus. Ser4 (blue) could not be assigned unambiguously.