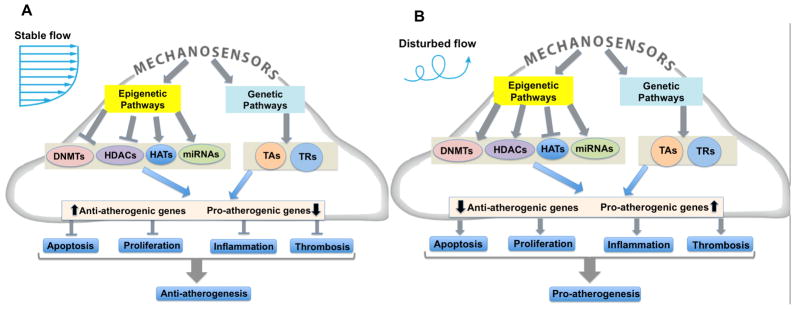
Figure 1. Flow regulates both genetic and epigenetic pathways involved in gene expression control – Working model.
(A) Stable flow (s-flow) downregulates DNA Methyltransferases (DNMTs) and histone deactylases (HDACs), and upregulates histone acetyltranferases (HATs). MicroRNAs (miRNAs) and transcription factors (both transcriptional activators (TAs) and transcriptional repressors (TRs) are also regulated by flow and demonstrate molecule-specific, coordinated responses. The interplay of these events leads to orchestrated transcription of a subset of anti-atherogenic genes and also prevents the expression of pro-atherogenic genes. (B) Under d-flow conditions, the opposite responses occur: HATs are downregulated and HDACs and DNMTs are overexpressed, leading to repression of a subset of anti-atherogenic genes and increased expression of a subset of pro-atherogenic genes. This mechanosensitive reprograming of gene expression by epigenomic DNA methylation leads to atherosclerosis development.