Figure 2.
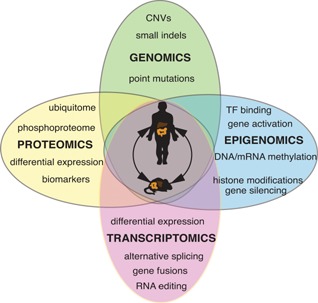
Cross‐species comparative ‘omics’ approaches for colorectal cancer (CRC) gene, therapeutic target and early disease biomarker discovery. Despite their evolutionary distance, the gene content of the mouse and human genomes has been largely conserved through evolution and most cancer pathways are operative in both species. Sporadic, low‐level mutagenesis of Apc and Kras in the mouse colon results in a high frequency of invasive adenocarcinomas and liver metastases. Furthermore, serial biopsies of patient tumours is not realistic, but with the development of mouse colonoscopes to guide biopsy collection, this is now a possibility in mouse models. Cancer gene, therapeutic target and biomarker discovery in patient specimens is challenging because of genetic, epigenetic and environmental heterogeneity between patients. Since many of these variables can be controlled for in the inbred laboratory mouse, the comparison of human and mouse primary and metastatic colorectal cancer genomes, transcriptomes, epigenomes, and proteomes including ubiquitinomes, will provide a powerful reductionist tool for identification of the molecular alterations that drive CRC invasion and metastasis. CNV, copy number variation; TF, transcription factor.
