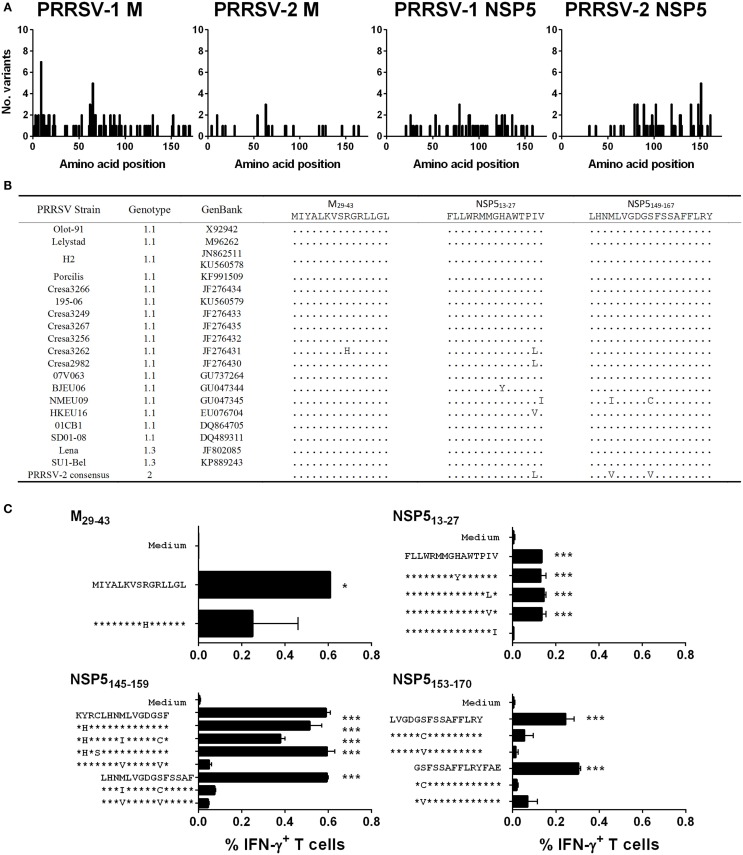
Figure 6.
Assessment of the conservation of identified T cell antigens M and NSP5 among PRRSV strains and assessment of T cell reactivity against variant peptides. The complete predicted amino acid sequences of M (from 64 PRRSV-1 and 31 PRRSV-2 strains) and NSP5 (from 19 PRRSV-1 and 36 PRRSV-2 strains) were aligned and the number of different amino acid variants at each residue plotted (A). The predicted amino acid sequences of identified antigenic regions, M29-43, NSP513-27, and NSP5149-167, were compared among the panel of 19 PRRSV-1 strains, for which both M and NSP5 sequence data were available, and a consensus sequence based on available PRRSV-2 strains (B). Based on the observed amino acid substitutions, variant peptides were used to stimulate PBMC from pigs 71 and 86 (C). IFN-γ expression by CD4 T cells to M29-43 (pig 71) and CD8 T cells to NSP513-27 (pig 71), NSP5145-159 (pig 86), and NSP5153-170 (pig 71) peptides were assessed by flow cytometry. The mean% of unstimulated (medium) and peptide stimulated data from duplicate cultures are presented and error bars show the SEM. Values were compared to the unstimulated control using a one-way ANOVA followed by a Dunnett’s multiple comparison test; ***p < 0.001, **p < 0.01, and *p < 0.05.