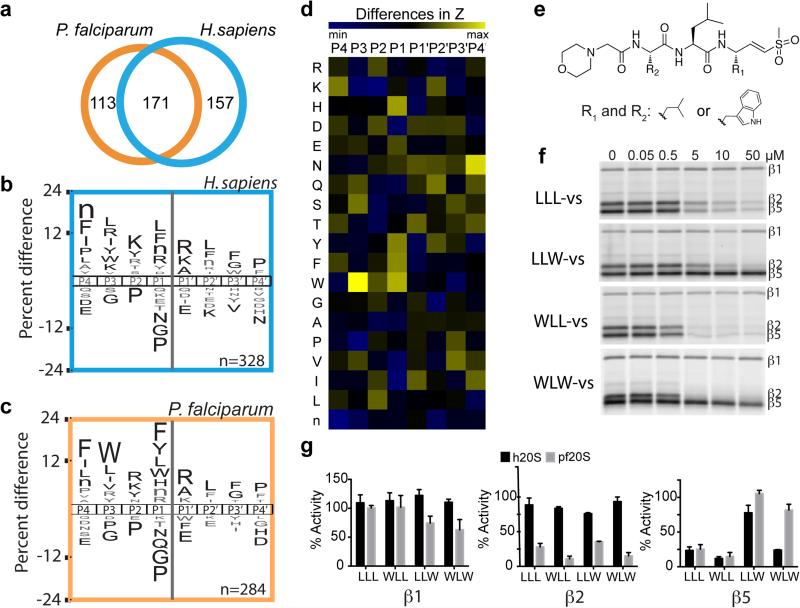
Figure 1.
Substrate profile of the activated human and P. falciparum 20S proteasome guides inhibitor design. a, Total number of cleavage sites detected after 4 hr incubation of the activated human and P. falciparum proteasome with the peptide library. The iceLogos generated from the cleavages are shown in (b) for human and (c) for P. falciparum proteasome. Amino acids that are most and least favored at each position are shown above and below the axis, respectively. Lowercase ‘n’ corresponds to norleucine and amino acids in black text are statistically significant (p < 0.05, unpaired two-tailed Student's t-test). d, The Z-score for amino acid at each position (P4-P4′) was calculated for both human and parasite proteasome based on the cleavages in a, and the difference between the Z-scores is shown as a heatmap. e, Inhibitors are designed by substituting Trp at either P1 and/or P3 position in the morpholino-capped tri-leucine vinyl sulfone. f, Inhibition of purified P. falciparum 20S as assessed by activity based probe labeling. The same experiment was repeated for the human 20S proteasome (Extended Data Figure 2b). g, Activity of each subunit in human or P. falciparum proteasome after 10 μM inhibitor treatment was determined by image quantification of the intensity of probe labeling and normalized to mock treated control. Error bars represent standard deviation (s.d.) and n=3 purified proteasome from 3 independent experiments (for gel source data, see Supplementary Fig. 1a and b).