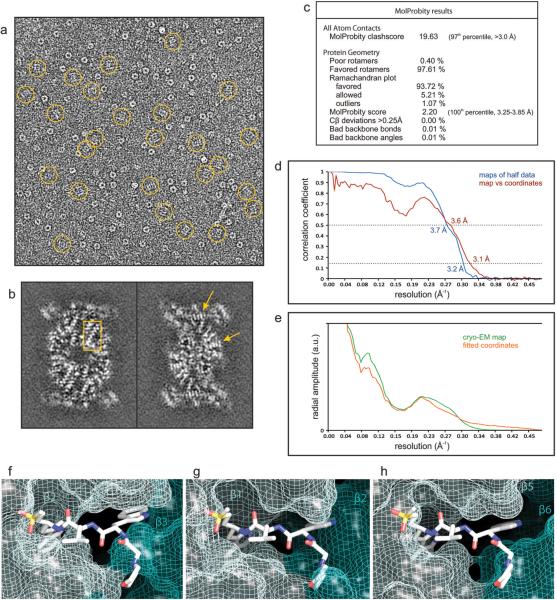
Extended Data Figure 3.
(a-e) Evaluation of the single particle analysis of the P. falciparum 20S proteasome core bound to the inhibitor WLW-vs. a, cryo-EM image of the sample analyzed, with molecular images of side views of the complex (normal to its long axis) indicated by rings. The image greyscale was inverted in order to show the protein densities in white. b, Individual sections of the 3D map, as determined by the 3D reconstruction algorithm (without further sharpening, masking or Fourier filtering), are represented as grey scale. These sections are 1 Å thick and reveal the quality of the reconstruction, as the protein densities are clearly resolved against a very smooth background, with regions showing the pattern of α-helices (box) and the clear separation of sheet forming β strands (arrows) indicated. c, Evaluation of the model of the P. falciparum 20S proteasome core using MolProbity2. d, Resolution estimate of the cryo-EM map by Fourier shell correlation. The curves correspond to the correlation obtained against the protein model (red) and the correlation between maps determined from two halves of the data (blue). The resolution was estimated from the curve against the model where the 0.5 correlation coefficient criterion3 yields an estimate of 3.6 Å. The correlation coefficient can be seen to fall to a local minimum at ~6 Å and then recover at higher resolutions for both FSC curves. This behavior is consistent with the rotationally averaged amplitude spectra of both the cryo-EM map and the coordinates (e). This region of the amplitude spectra contains reduced structural information, typical of protein scattering, indicating that these effects in the FSC curves arise from a genuine local reduction in the signal:noise ratio. (f-h) Accessibility of the human 20S proteasome active sites to the inhibitor WLW-vs, using the protein model of the human proteasome core complex bound to a LLL-vs inhibitor4. Protein coordinates of the human proteasome 20S core (PDB accession code 5a0q) β2 (f), β1 (g) and β5 (h) active sites were aligned to the coordinates of the P. falciparum proteasome β2 subunit bound to the WLW-vs inhibitor. The model of the human 20S proteasome active sites is represented as van der Waals surfaces with the superimposed WLW-vs inhibitor shown as sticks.