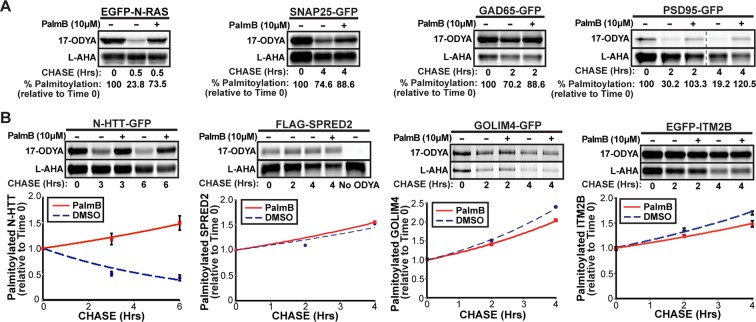
Figure 1. Dual-click chemistry labeling reveals differences in protein depalmitoylation dynamics.
(A) Pulse-chase analysis of established palmitoyl-proteins (N-Ras, SNAP25, GAD65, PSD95) by dual-click chemistry in the presence of DMSO (-) or 10 μM PalmB (+). Representative in-gel fluorescence scans illustrate dual detection of 17-ODYA (palmitate analogue) and L-AHA (methionine analogue) using Alexa Fluor 488 and Alexa Fluor 647, respectively. Dashed line indicates cropping of a single gel. n = 2 per substrate. (B) Pulse-chase analysis of palmitate turnover on N-HTT, SPRED2, GOLIM4, and ITM2B by dual-click chemistry as described in (A). Upper panels: representative in-gel fluorescence scans; Lower panels: Time course of substrate depalmitoyation in DMSO- and PalmB-treated cells after normalizing 17-ODYA to L-AHA signals at each chase time. n = 2, mean ± SEM. 17-ODYA, 17-octadecynoic acid; L-AHA, L-azidohomoalanine; SEM, standard error of the mean.