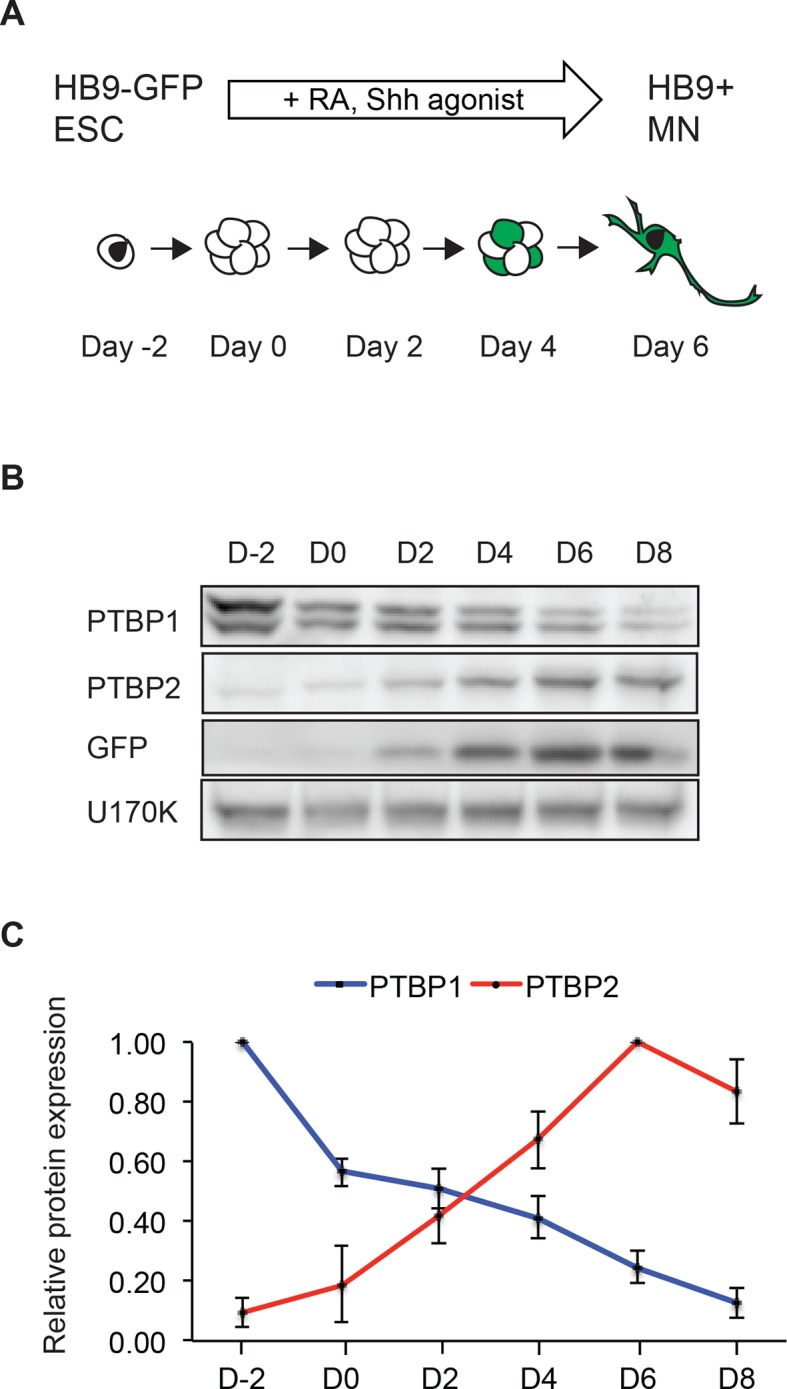
Figure 1. The transition in PTB protein expression occurs during in vitro neuronal differentiation.
(A) HB9-GFP ESCs express eGFP under the MN-specific promoter HB9. The addition of retinoic acid (RA) and a Sonic hedgehog (Shh) agonist drives MN differentiation. (B) Western blot shows the loss of PTBP1 protein and gain of PTBP2 and GFP proteins as ESCs differentiate into HB9+ MNs. U170K served as a loading control. (C) Quantification of relative PTBP1 and PTBP2 protein expression across MN differentiation. Error bars represent standard error of the mean (SEM, n=2).
DOI: http://dx.doi.org/10.7554/eLife.09268.003