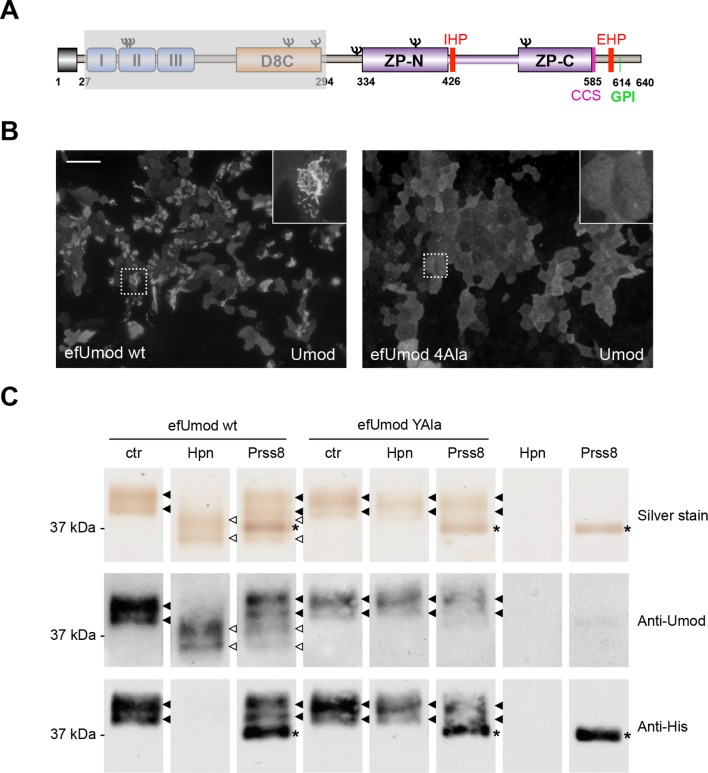
Figure 4. Hepsin and prostasin directly cleave uromodulin in vitro.
(A) Schematic representation of human uromodulin domain structure as shown in Figure 1A. The region not included in recombinant efUmod is shadowed. (B) The deletion of the elastase-sensitive fragment of uromodulin does not affect protein polymerisation on the surface of MDCK cells, as shown by immunofluorescence analysis (efUmod wt). As for full-length uromodulin (Schaeffer et al., 2009), this process depends on correct protein cleavage at the physiological site, since it is abolished when the consensus cleavage site is mutated (efUmod 4Ala, carrying the mutation 586RFRS589 > 586AAAA589). Scale bar, 50 µm. (C) Purified efUmod, either wild-type (efUmod wt) or mutated at the consensus cleavage site (efUmod YAla, carrying the mutation 586RFRS589 > 586AYAA589), was incubated with recombinant prostasin or hepsin, as indicated. Both proteases decrease the mass of wild-type efUmod (white arrowheads in upper and middle panels) and cause the loss of its C-terminal His-tag (lower panel). Hepsin is more efficient than prostasin, as it drives complete digestion of the product, despite being used at 20x lower concentration (picomolar ratio between protease and efUmod was 1:100 for hepsin and 1:5 for prostasin, see lanes 7 and 8 for comparison). The asterisk indicates His-tagged prostasin.