Figure 5. CIDEA is a phosphatidic acid (PA)-binding protein.
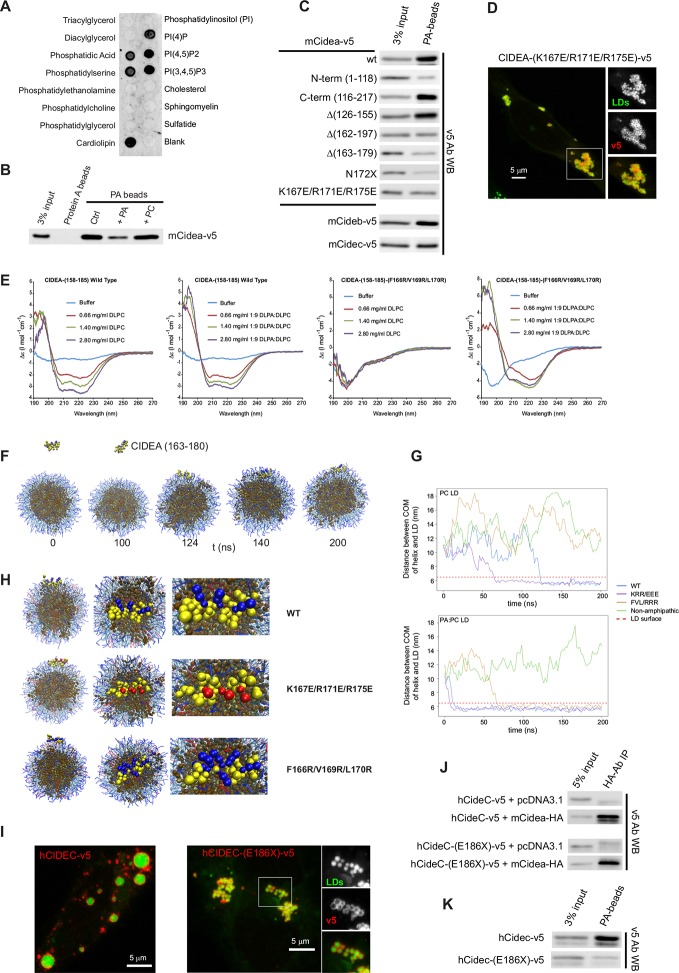
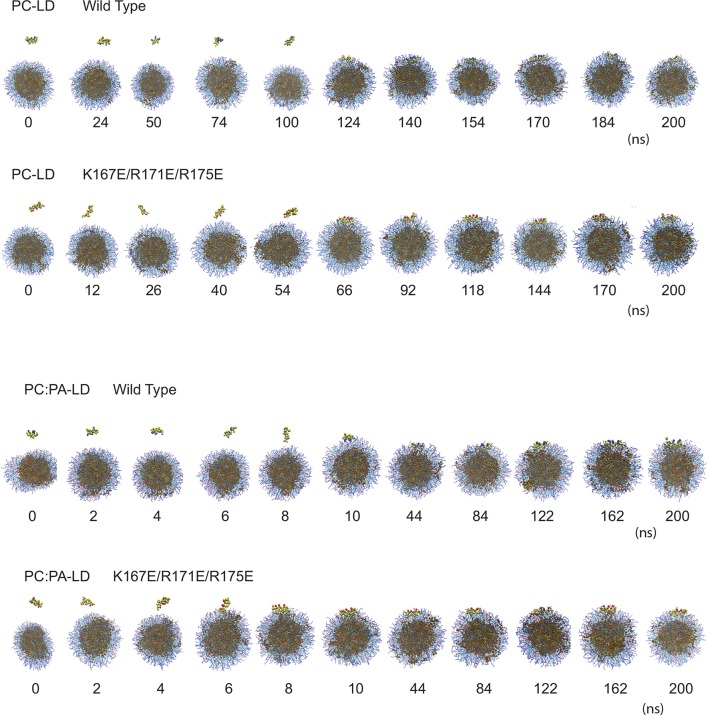
(A) Lipid strip assay showing the affinity of CIDEA-v5 for certain anionic phospholipids. (B) Interaction of CIDEA-v5 with PA beads. Binding was reduced by pre-incubation of the lysate with soluble PA, but not phosphatidylcholine (PC). (C) The affinity for PA beads was highly reduced in CIDEA-v5 constructs with alterations in its C-term hydrophobic and basic region (162–197). (D) CIDEA-(K167E/R171E/R175E)-v5 localizes to LDs and induces their clustering but cannot promote their enlargement by lipid transfer. Representative images are shown of experiments performed in a minimum of three independent experiments for every construct (n>50 cells). (E) Circular dichroism spectra of the synthetic wild type (wt) or mutant (F166R/V169R/L170R) CIDEA peptides encompassing aas 158–185 solubilized in 25 mM sodium phosphate (pH 7.2) at concentrations of 70 μM (wt) and 47 μM (mutant). Peptide samples were prepared in the absence and presence of increasing amounts of DPLC or DLPC:DLPA (9:1 molar ratio). (F–H) Coarse-grained molecular dynamics (CG-MD) simulations of peptide interactions with LDs (PC: hydrophobic chains, transparent blue, polar heads, opaque blue; TAG: hydrophobic chains, dark brown, glycerol chain, light brown; PA: hydrophobic chains, transparent red, polar heads, opaque red; peptides: yellow, with cationic aa in blue and anionic in red). (F) Selected time points of the wt helix simulation with PC-LDs. At 124 ns the helix initiates the contact through its hydrophobic face, being rapidly embedded in the phospholipid monolayer. TAG molecules can abandon the neutral lipid core and are integrated in the hydrophobic region of the phospholipid monolayer. (G) Distance between the peptide and LD centre of mass (COM) versus time for the different helices with a PC-LD and a PC:PA-LD. The dashed line represents the approximate location of LD phospholipid head groups. (H) Different views of the configuration of the LD helix at the end of the simulations. Interaction between the polar head of PA and the helix can be observed for the wt and F166R/V169R/L170R but not K167E/R171E/R175E. (I–K) Comparison of full-length hCIDEC-v5 and the lipodystrophy-associated truncation hCIDEC-(E186X)-v5, including LD localization and morphology (I), co-IP with CIDEA-HA (J), and affinity for PA beads (K). Each co-IP, PA-binding assay, and lipid strip assay was performed at least in triplicate, producing similar results in each experiment.
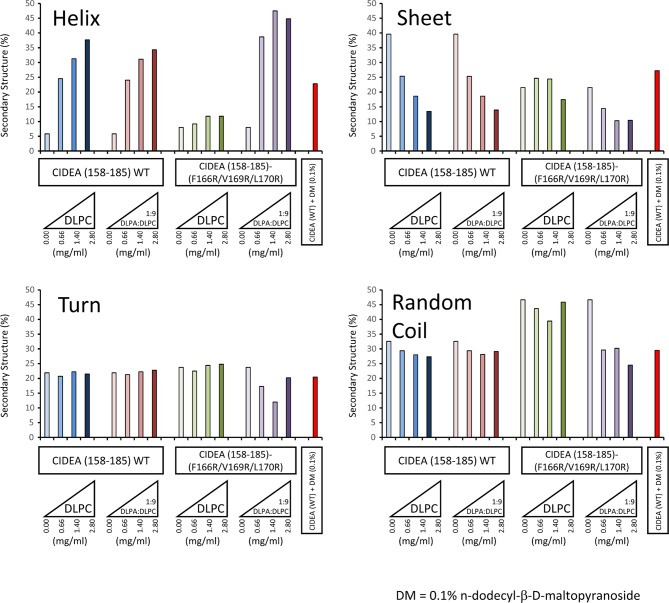
Figure 5—figure supplement 1. Secondary structure determination of CIDEA amino acids 158–185 (wild type and F166R/V169R/L170R) by CDPro DATABASE 4 (43 soluble proteins) using the CONTINLL program.
Figure 5—figure supplement 2. Computational prediction of the amphipathic helix and LD interactions.
Figure 5—figure supplement 3. TAG infiltration into the phospholipid monolayer.
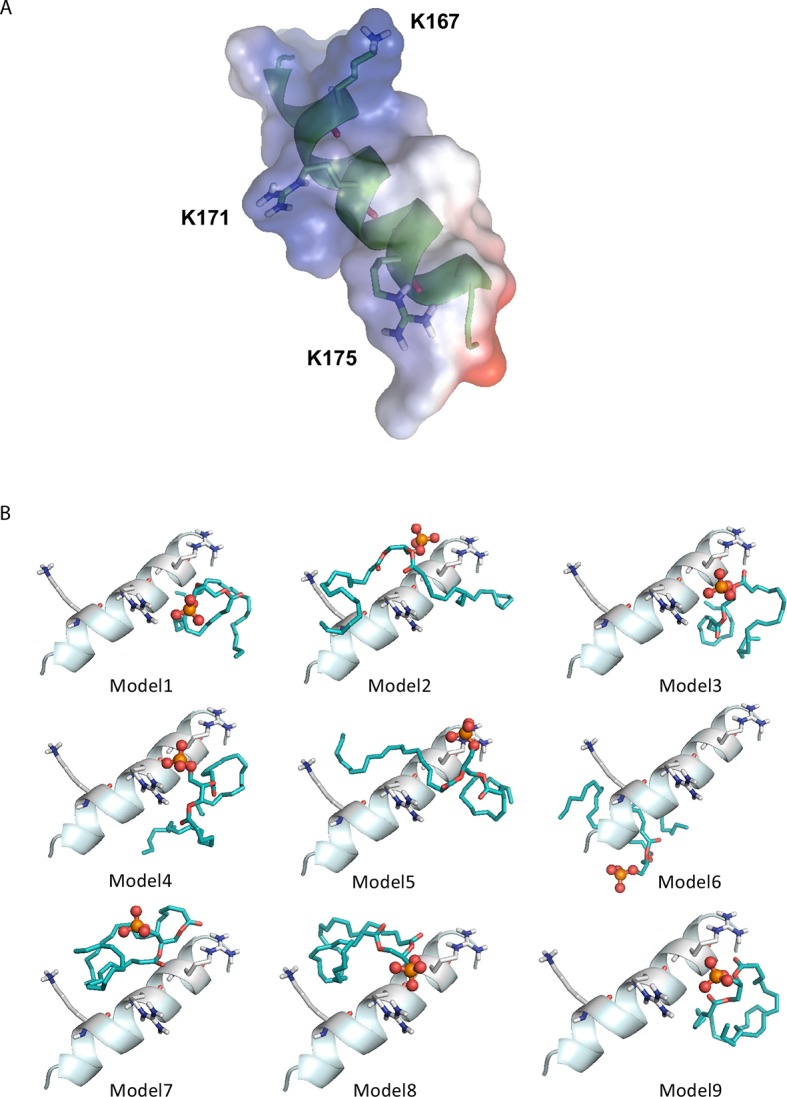
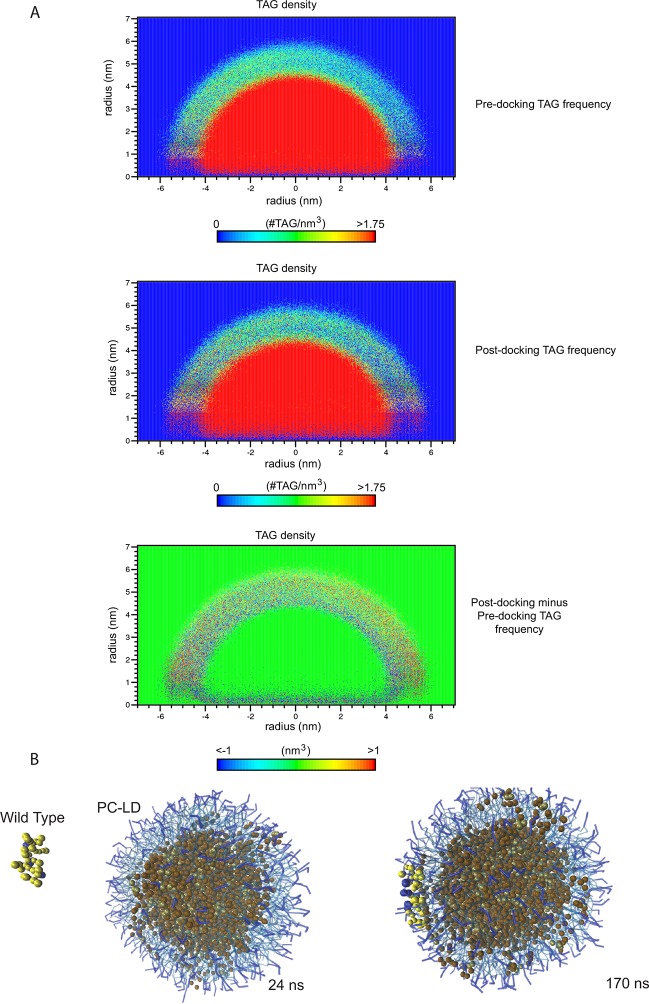
Figure 5—figure supplement 4. Computational prediction of PA docking to the amphipathic helix structure of CIDEA.