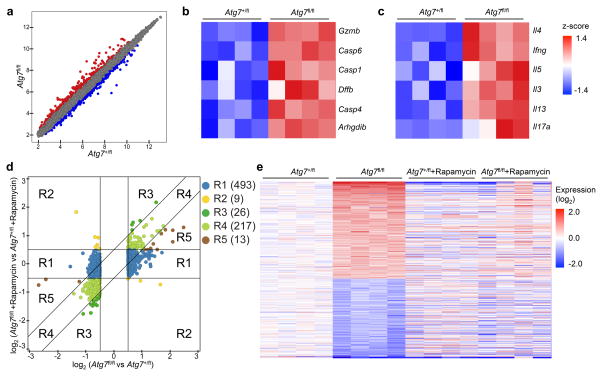
Figure 4. mTORC1 signaling is critical for Atg7-dependent transcriptional program.
Foxp3CreAtg7+/fl and Foxp3CreAtg7fl/fl mice were treated with or without rapamycin (n=4 mice per group). Treg cells were sorted and activated with anti-CD3 and anti-CD28 for 4 h for gene expression profiling. (a) Scatterplot comparing global gene-expression profiles between Foxp3CreAtg7+/fl and Foxp3CreAtg7fl/fl Treg cells. Transcripts with > 0.5 log2 fold increased (360 probes) or decreased (398 probes) expression in Foxp3CreAtg7fl/fl Treg cells are shown in red or blue respectively. (b,c) GSEA reveals that caspase (b) and cytokine pathways (c) are among the most extensively upregulated pathways in Foxp3CreAtg7fl/fl Treg cells as compared with Foxp3CreAtg7+/fl Treg cells. Heat maps of top hit genes in caspase (b) and cytokine (c) pathways. Differentially expressed genes were normalized by z-score. Expression levels are shown as green for low intensities, and red for high intensities. (d) Comparison of expression changes in rapamycin-treated Foxp3CreAtg7fl/fl versus Foxp3CreAtg7+/fl Treg cells with those in non-treated Foxp3CreAtg7fl/fl versus Foxp3CreAtg7+/fl Treg cells. The 758 Atg7 target genes (with > 0.5 log2 fold change) were partitioned into five main clusters, shown and colored by regions (R1–R5). Right, numbers indicate the number of probes within each region. (e) Heat maps of 515 rapamycin responsive genes that are differentially expressed in Foxp3CreAtg7fl/fl Treg cells (with > 0.5 log2 fold change) and have diminished response after rapamycin treatment. Red color denotes upregulated genes in Foxp3CreAtg7fl/fl Treg cells, and blue color denotes downregulated genes in Foxp3CreAtg7fl/fl Treg cells. Data are from one experiment (a–e).