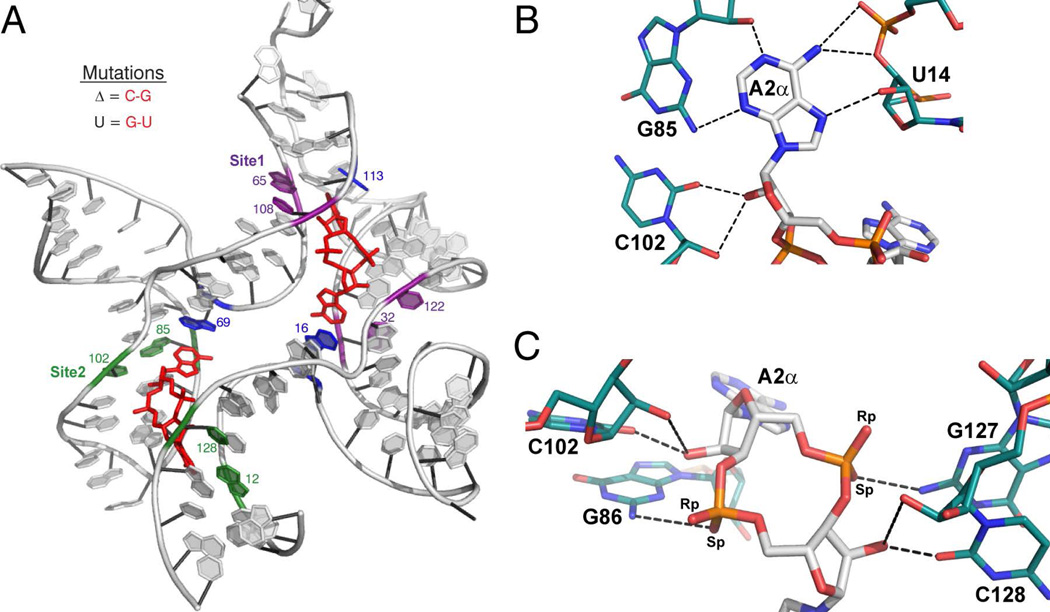
Figure 2.
c-di-AMP recognition by the ydaO riboswitch (ref. 45–47). (A) Structure of the ydaO aptamer from B. subtilis bound to two molecules of c-di-AMP. c-di-AMP is colored in red. Conserved Watson-Crick G-C pairs involved in type-I A-minor interactions with c-di-AMP are shown in purple for Site 1 and green for Site 2. Bases that stack with the adenines are shown in blue. Important nucleotides are labeled. (B and C) c-di-AMP-A2α recognition by the ydaO motif. c-di-AMP is colored by atom with carbon in white, oxygen in red, nitrogen in blue, and phosphorous in orange. Hydrogen-bonding nucleotides of ydaO are colored as described, with carbons in teal. Hydrogen bonds are shown as black dashed lines. (B) Recognition of the Watson-Crick and Hoogsteen faces of the c-di-AMP nucleobases featuring the type-1 A-minor interaction. (C) Backbone recognition of c-di-AMP by the ydaO riboswitch. These c-di-AMP recognition motifs are observed with each adenosine of the ligand.