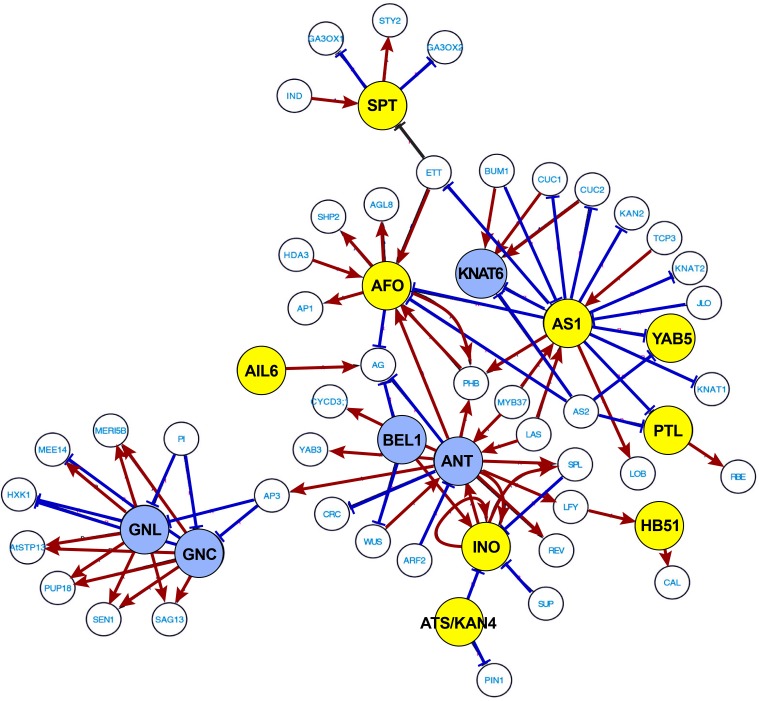
Figure 5.
Most extensive transcriptional network underlying soybean leaf abscission. The 188 abscission-specific soybean TFs corresponding to 133 Arabidopsis TFs (Table S2) were used to construct high-confidence transcriptional networks using the Arabidopsis Transcriptional Regulatory Map (ATRM). Of 133 Arabidopsis TF homologs, only 40 of Arabidopsis TFs were found in the ATRM data set (Table 1, Table S5). Arabidopsis TF orthologs identified in expression data of soybean leaf abscission were color-coded. Soybean TFs eight-fold higher in the LAZ/NAZ are highlighted in yellow and TFs eight-fold lower in the LAZ/NAZ are highlighted in blue. Visualization of the network was generated by Cytoscape v. 3.2.1. Red arrows indicated a positive regulation and blue bars a negative regulation of the target TFs or co-regulators generated from ATRM data set.