Abstract
Genes which confer partial resistance to the rusts in wheat figure prominently in discussions of potential durable resistance strategies. The positional cloning of the first of these genes, Lr34/Yr18 and Yr36, has revealed different protein structures, suggesting that the category of partial resistance genes, as defined by phenotype, likely groups together suites of functionally heterogenous genes. With the number of mapped partial rust resistance genes increasing rapidly as a result of ongoing advances in marker and sequencing technologies, breeding programs needing to select and prioritize genes for deployment confront a fundamental question: which genes or gene combinations are more likely to provide durable protection against these evolving pathogens? We argue that a refined classification of partial rust resistance genes is required to start answering this question, one based not merely on disease phenotype but also on gene cloning, molecular functional characterization, and interactions with other host and pathogen proteins. Combined with accurate and detailed disease phenotyping and standard genetic studies, an integrated wheat-rust interactome promises to provide the basis for a functional classification of partial resistance genes and thus a conceptual framework for their rational deployment.
Keywords: Puccinia, Triticum aestivum, Partial resistance, Interactome, Breeding
Background
The cereal rusts (Puccinia spp.), caused by highly specialized Basidiomycetes within the order Uredinales, are among the most destructive plant diseases, responsible for recurrent episodes of catastrophic yield loss and economic hardship among grain-based agricultural societies for centuries. Within this broad class of diseases, stem rust (SR: causal organism P. graminis Pers. f. sp. tritici), stripe rust (YR: causal organism P. striiformis f. sp. tritici), and leaf rust (LR: causal organism P. triticina) of wheat (Triticum aestivum) are prominent in their historic relevance to agricultural productivity. While it is possible to mitigate losses to these pathogens through the timely application of fungicides, the low profitability of wheat production cannot sustain such costly inputs under many dryland situations. Consequently, emphasis has long been placed on the identification and deployment of genetic sources of resistance.
Major resistance genes
Most of the resistance genes discovered and deployed in defense against the wheat rusts are classified as major resistance genes. Also known as seedling resistance, vertical resistance, all-stage resistance, or race-specific resistance genes, major genes confer effective levels of resistance against specific physiologic races of the pathogen, generally throughout the life cycle of the host. In terms of disease phenotype, major resistance genes are often associated with a rapid programmed death of infected cells, a so-called “hypersensitive response” thought to play a role in limiting the expansion of biotrophic pathogens in the host tissue. Major resistance genes have been systematically investigated since the first such genes (e.g. Pto from tomato and RPS2 from A. thaliana) were cloned in the early 1990’s (Martin et al. 1993; Bent et al. 1994; Mindrinos et al. 1994). In the monocots, nine genes conferring resistance to Puccinia spp. have been cloned, with most exhibiting the form of intracellular receptors with a common NBS-LRR structure (nucleotide binding site + leucine rich repeat domain) (see Table 1). The proposed mechanisms of race-specificity and the actions of such major genes, including R-Avr gene interactions and the guard hypothesis, have been discussed in detail in previous papers and reviews (e.g. Hulbert et al. 2001; Jones and Dangl 2006; Bent and Mackey 2007).
Table 1.
The structures of known genes conferring resistance to Puccinia spp. in cereals (Cloutier et al. 2007; Brueggeman et al. 2008; Fu et al. 2009; Krattinger et al. 2009; updated from Ayliffe and Lagudah 2004). Despite the general trend presented here, lack of durability is not exclusively or even necessarily associated with the NBS-LRR forms of resistance genes, as illustrated by the emergence of pathotype Pgt-QCC virulent on the long-deployed barley stem rust resistance gene Rpg1 (a protein kinase) (Martens et al. 1989; Jin et al. 1994; Brueggeman et al. 2002) and the current efficacy of the leaf rust resistance gene Lr21 (an NBS-LRR gene of limited deployment) against all known races of leaf rust (Huang et al. 2009)
| Major resistance genes with known or predicted race specificity | |||
|---|---|---|---|
|
| |||
| Species | Gene | Putative gene product | Disease |
| Wheat | Lr1 | CC-NBS-LRR | Leaf rust |
| Lr10 | NBS-LRR | Leaf rust | |
| Lr21 | NBS-LRR | Leaf rust | |
| Maize | Rp1-D | NBS-LRR | Leaf rust |
| Rp3 | NBS-LRR | Leaf rust | |
| Barley | Rpg1 | Kinase-kinase | Stem rust |
| Rpg5 | NBS-LRR-Kinase | Stem rust | |
|
| |||
| Partial resistance genes with possible race non-specificity | |||
|
| |||
| Wheat | Lr34/Yr18/Pm38 | ABC transporter | Leaf rust/stripe rust/powdery mildew |
| Yr36 | Kinase-START | Stripe rust | |
The traditional emphasis on major gene resistance is understandable for many reasons:
Major genes are effective: Such genes have, without question, provided significant economic benefits to wheat growers, as illustrated by the decades-long protection provided by the now-defeated stem rust resistance gene Sr31;
They are relatively easy to identify and deploy: Breeding for single-gene resistance is much simpler than breeding for oligo/polygenic resistance;
They are historically easier to combine: In the absence of clear additivity of the resistance conferred by different genes and in the absence of good molecular markers, race-specific disease phenotypes facilitate the efficient introgression and stacking of major genes using panels of different rust races
They provide clear levels of protection: “Resistance,” rather than “tolerance”, has long been held as an agronomic goal.
Despite these clear historic advantages, however, major resistance genes frequently lacked “durability”, that is, the ability of a widely-deployed resistance gene to provide an economic level of protection over an extended period of time (Johnson 1984). To illustrate this fact, consider that of the approximately 70 identified major genes for resistance to stripe rust, only three (Yr5, Yr15, and Yr24/26, each neither widely nor long-deployed) remain effective against all detected races of the pathogen in the United States (Chen 2007). With the Ug99 family of stem rust races diversifying (Jin et al. 2008, 2009)1, and with nearly 80 races of stripe rust with new virulence patterns appearing in the United States alone since 20002, it is generally agreed that the defeat of major genes is no longer a question of “if” but “when” and that the need for alternative defense strategies is pressing. Specifically, these recent epidemics have reinvigorated interest within the wheat breeding and research communities in partial resistance genes as sources of potentially more durable resistance.
Partial resistance genes
Genes which confer partial resistance to the cereal rusts are variously referred to as race non-specific, (high-temperature) adult plant, horizontal, or slow-rusting resistance genes. Generally speaking, whichever term is used, partial resistance is understood to mean a form of resistance that is not complete (i.e. all stages of the infection process can occur, including sporulation), is not associated with a rapid hypersensitive response, is not equally effective throughout all stages of host development (e.g. it is often undetectable at the seedling stage), and is not known to be limited to specific physiologic races of the pathogen. In other words, partial resistance is a catch-all designation which has come to mean any form of resistance that is not “major”. As such, the term likely groups together disparate classes of functionally heterogenous resistance genes and provides very little information regarding their functional relatedness, combinability, or potential durability.
The recent cloning of the first two partial rust resistance genes in wheat, namely the pleiotropic leaf rust/stripe rust/powdery mildew resistance gene Lr34/Yr18/Pm38 (Krattinger et al. 2009) and the stripe rust resistance gene Yr36 (Fu et al. 2009), provides support to the hypothesis that partial rust resistance genes comprise a heterogeneous group of functionally diverse genes. The historically durable resistance gene Lr34/Yr18/Pm38 encodes a protein with homology to ATP-binding cassette (ABC) transporters, a transkingdom superfamily of transmembrane proteins involved in the transport of a wide variety of substrates across cellular membranes (Krattinger et al. 2009). In addition to its structure (non-NBS-LRR) and longstanding durability (more than 50 years), other pieces of evidence suggest that the partial resistance conferred by Lr34/Yr18 is truly race non-specific. To begin, its pleiotropic effect against leaf rust, stripe rust, and powdery mildew (another fungal disease caused by the biotroph Blumeria graminis) indicates that the gene is in fact species non-specific. The gene is also associated, even in non-challenged tissue, with leaf-tip necrosis (Singh 1992), a constitutive physiological characteristic at odds with the needs of biotrophic pathogens.
The other recently-cloned slow-rusting gene, conferring partial resistance to wheat stripe rust, is the high-temperature adult plant (HTAP) gene Yr36. Based on its sequence, this gene encodes a protein with a kinase domain linked to a steroidogenic acute regulatory protein-related lipid transfer (START) domain predicted to bind lipids. The novelty of this protein architecture is evidenced by the fact that these two domains are not found together in any protein from any organism currently deposited in GenBank. While race non-specificity for Yr36 is suggested based on the resistance it confers against the few broadly-virulent pathotypes tested to date (Fu et al. 2009), claims about the durability and race non-specificity of Yr36 will need to wait for its wider and longer deployment in commercial varieties.
The gene-for-gene hypothesis of host-pathogen compatibility (Flor 1942, 1956), inspired by and relevant to the major class of race-specific NBS-LRR genes, provided an appropriate conceptual framework for the rust research community for many decades. But as the focus of research expands to encompass non-NBS-LRR sources of partial resistance and the potential achievement of more durable resistance through their strategic deployment, a new conceptual framework is required. Assuming the first two cloned partial resistance genes are any indication of the diversity of genes involved, such a new framework will be neither as simple nor as predictive of the protein structures of resistance genes as the gene-for-gene model. Fortunately, technologies more truly commensurate with the complexity of the Triticum-Puccinia interactions are becoming available, raising the possibility of a more meaningful classification of non-major resistance genes.
The quest for more durable resistance
While durability is not necessarily associated with partial resistance, as illustrated by the emergence of virulent P. striiformis tritici races on the partially resistant varieties Joss Cambier, Maris Bilbo, and others (Johnson 1978), several partial resistance genes have historically played central roles in durable resistance strategies. Examples of the longstanding effectiveness of combinations of partial resistance genes include the cv. Stephens-type resistance to stripe rust (Santra et al. 2008), the protection provided by the slow-rusting Sr2 complex to stem rust (Singh et al. 2006), and the widespread combination of Lr13 and Lr34 for leaf rust (Kolmer 1996). Other combinations of partial resistance genes in the CIMMYT breeding program have conferred adequate resistance to leaf rust in the field for several years (Singh et al. 2000). Indeed, the potential importance of partial resistance genes in achieving durable protection against the cereal rusts has gained great traction since its first observation over 50 years ago (Caldwell et al. 1958; Johnson 1981) and today serves as a rough guiding principle to many research programs.
This renewed interest in partial resistance has accelerated the discovery rate of partial resistance QTL. In the last two years alone, more than 20 QTL conferring some degree of quantitative resistance to wheat stripe rust were reported (Cheng and Chen 2008; Chhuneja et al. 2008; Guo et al. 2008; Melichar et al. 2008; Rosewarne et al. 2008; Santra et al. 2008; Carter et al. 2009; Dedryver et al. 2009; Lu et al. 2009). These QTL present breeders with the possibility of over 1,000 unique three-QTL combinations, a number far beyond the realm of investigation. With the development of ever more precise and affordable molecular markers, it is also possible to deploy combinations of partial and major resistance genes, further increasing the number of available breeding options. How should a breeding program, constrained by limited resources, respond to such data overload? To answer this question, a means of predicting the relative durability of combinations of resistance genes is needed. In this paper, we argue that a research program capable of achieving this objective will seek to answer the following basic questions:
Can partial resistance genes be assigned to functional classes predictive of their contributions to overall resistance in resistance gene combinations?
Are combinations of resistance genes from different functional classes more or less durable than combinations of genes from the same functional class?
A systems biology approach
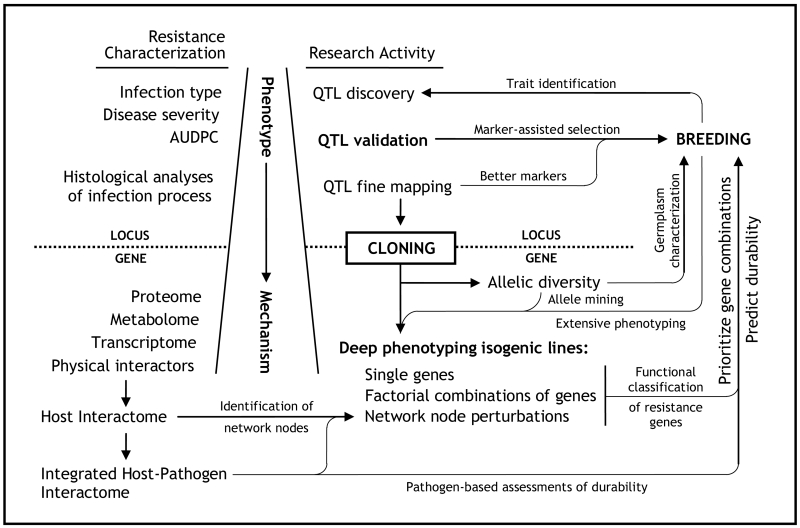
Research on model organisms, from E. coli to A. thaliana to S. cerevisiae, has revealed certain fundamental properties of biological systems worth noting (Ideker et al. 2001; Kitano 2002; Mustacchi et al. 2006; Van Norman and Benfey 2009). First and foremost among these is this guiding principle from the field of systems biology: Biological systems are more accurately modeled as adaptive, environmentally-responsive networks of highly connected nodes rather than as collections of rigidly-defined biochemical pathways populated by dedicated, single-function gene products (Benfey and Mitchell-Olds 2008). One implication of this insight is that far more will be required than the phenotypic, or even genetic, characterization of individual resistance genes if we are ever to understand, and thus conceivably engineer, reliably durable resistance to the wheat rusts. As expounded in detail throughout the remainder of this paper, a more integrated, systems-oriented approach to these host-pathogen systems is necessary to generate a meaningful functional classification of partial resistance genes and thus provide a much needed conceptual framework for their deployment (Fig. 1).
Fig. 1.
Cloning genes underlying broadly effective partial resistance loci is the required point of entry into a research program dedicated to the functional characterization of rust resistance genes in wheat. Cloning permits the rapid assessment of allelic diversity in both research and breeding materials and is thus necessary for the thorough identification, evaluation, and utilization of natural sources of resistance. Cloning also opens the possibility of more extensive and unbiased phenotypic characterizations which take advantage of high-throughput molecular platforms. Once genes are known, their encoded proteins can be placed within interaction networks to which strategic perturbations may be applied, thereby facilitating their functional characterization. Expansion of the interactome to include pathogen proteins offers an additional means of identifying network perturbation targets and assessing the likelihood of true race non-specificity. Integrated with classical phenotypes and rigorous epistatic studies, such information can help prioritize gene combinations for breeding and is likely our best chance to predict and engineer durable resistance. Breeding programs benefit from and are in a position to assist such basic research at all stages, both short- and long-term. (QTL: quantitative trait locus; AUDPC: area under the disease progress curve)
The necessary first step: cloning partial resistance genes
Cloning partial resistance genes is the minimum requisite entry-point into a systems-biology oriented research program dedicated to the functional classification of partial resistance genes; thus we assert that a short-term objective of the rust research community should be to build a catalog of cloned host genes associated with partial resistance under field conditions. Cloning genes and determining the allele(s) responsible for the resistance associated with a given genetic locus provides valuable information at multiple levels, making possible:
The more efficient introgression of a gene into multiple backgrounds using the allele itself as a diagnostic (perfect or functional) marker in MAS;
The systematic characterization of allelic diversity in germplasm collections to identify other functional resistance alleles (allele mining);
The efficient screening of breeding lines and varieties to determine the presence and frequency of resistance and susceptibility alleles in breeding programs;
A functional characterization of the gene based on its expression and translation profiles, its cellular localization, and the molecular interactions of its protein product;
The use of transgenic methods to accelerate the development of experimental isogenic lines (for environmental and epistatic studies), to develop insertable single-locus cassettes of multiple resistance genes, and to test the phenotypic and molecular effects of gene over-expression and down-regulation.
Fortunately, numerous technologies are converging to significantly decrease the cost of cloning genes in wheat. Large collections of mapped expressed sequence tags (ESTs), single nucleotide polymorphisms (SNPs), restriction fragment length polymorphisms (RFLPs), and microsatellites (SSRs) make the wheat genome one of the most densely-mapped among those yet to be sequenced fully (Keller et al. 2005). The sequences of these mapped markers serve as anchors to the full reference genomes for rice (International Rice Genome Sequencing Project 2005) and Brachypodium (International Brachypodium Initiative 2010), making possible the relatively rapid construction of high-density maps by enabling, via comparative genomics, the development of molecular markers targeted to specific genomic regions. The construction of local physical maps is further facilitated by the availability of reference physical maps of Aegilops tauschii (wheatdb.ucdavis.edu) and some T. aestivum chromosomes (Paux et al. 2008) as well as by major advances in sequencing technologies (for a review, see Metzker 2010). By generating DNA sequence data in a massively parallel fashion at substantially lower cost than traditional Sanger sequencing approaches, next generation sequencing (NGS) platforms greatly simplify the sequencing of local wheat genomic regions and thus substantially reduce the cost of map-based cloning in wheat. These NGS platforms also accelerate efforts to sequence the complete wheat genome, as evidenced by the recent availability of a 5× 454 sequence of the reference genotype Chinese Spring (www.cerealsdb.uk.net/search_reads.htm).
Beyond disease phenotype, toward a functional classification of resistance genes
By necessity, the early rust researchers and breeders relied upon macroscopic host phenotype as the single lens through which to view host-pathogen interactions and to guide breeding strategies. Even today, the visible disease response of differential host lines serves as the primary means by which pathogen variability is investigated, the “durability” of deployed major resistance genes is monitored, and the potential utility of major gene combinations, given current pathotypes, is postulated. Once a partial resistance gene is cloned, however, it becomes possible to view and characterize host-pathogen interactions in new and powerful ways. For example, mutants or RNAi transgenic lines with altered transcript levels (see review by Fu et al. 2007) can be developed to investigate the function, not just the associated phenotype, of a cloned resistance gene. Through transcriptome (using NGS), proteome, and metabolome comparisons of wild type and mutant or RNAi transgenic lines, cloned partial resistance genes may begin to be classified according to which defense responses and metabolic pathways they activate (Eulgem 2005; Glazebrook 2005), whether they induce or inhibit programmed cell death (Tsunezuka et al. 2005), and the chemical nature of their associated secondary metabolites (Fiehn 2002). As a complement to these molecular strategies, traditional histological analyses of fungal structures at different time points after inoculation continue to offer a valuable means of discriminating resistance genes based upon the stages at which they are observed to impair disease development. For example, while Lr34/Yr18 is reported to condition prehaustorial resistance (Rubiales and Niks 1995), Yr36 greatly restricts the post-haustorial spread of the fungus through the host tissue and is associated with the increased presence of autofluoresecent mesophyll cells (Fu et al. 2009). Ultimately, a meaningful classification of partial resistance genes will make use of all available descriptive information, from classical disease phenotype to traditional histological characterization to the numerous forms of molecular profiling made possible only once such genes are cloned.
Cloned genes as entry-points to defense-related gene networks
Non-covalent interactions among proteins are fundamental to all biological processes, including defense response signaling (Morsy et al. 2008). Determining how a resistance protein interfaces with protein-protein interaction networks (i.e. regions or neighborhoods within the host’s interactome, its complete universe of interacting proteins) can greatly improve our understanding of its biological functions and further facilitate the functional characterization of the gene by linking it physically to different signal transduction or metabolic pathways that contribute to the partial resistance phenotype. By placing resistance genes within their associated gene networks, new regulators of defense responses that are effective against rust pathogens may be identified.
The rice defense interactome has already proven to be a powerful tool for dissecting the signal transduction pathway of Xa21, a defense gene which confers resistance to bacterial leaf blight of rice (Song et al. 1995). By extending the interactome network centered on Xa21, new defense response proteins have been identified, revealing insights into the signal transduction pathways governing disease resistance (Wang et al. 2006; Park et al. 2008; Peng et al. 2008). Interactome network maps can be developed using high-throughput protein-protein interaction studies and then validated by multiple orthogonal in vivo and in planta interaction methods (Cusick et al. 2005; Morsy et al. 2008). For example, large-scale screening for interacting proteins and the construction of protein interactomes can be accomplished using a yeast-two-hybrid screen of whole cDNA libraries (Ding et al. 2009) or via large-scale mass spectrometry of tandem affinity purified-tagged proteins (Rohila et al. 2006). In wheat, yeast-two-hybrid libraries have been developed from stripe rust-infected leaves and are now being used to construct the local Yr36 interactome (collaboration between J. Dubcovsky and P. Ronald laboratories).
Defining the interactome neighborhoods around cloned resistance genes may make it possible to identify local network hubs (or nodes) that, in a very direct sense, could serve as bases for the mechanistic classification of resistance genes (i.e. functional relatedness). For example, the trimeric molecular chaperone HSP90-SGT1-RAR1 physically interacts with and is known to be required for the resistance mediated by many NBS-LRR resistance genes such as Mla6 and Mla12, effective against powdery mildew in barley (Zhou et al. 2001; Azevedo et al. 2002), and Lr21, effective against leaf rust in wheat (Scofield et al. 2005; Seo et al. 2008; Shirasu 2009). Phenotypic and molecular characterizations of isogenic lines (with and without the target resistance gene) under systematic perturbations or disruptions of the HSP90-SGT1-RAR1 complex, a known interactome network node, could permit the characterization of resistance genes as either HSP90-SGT1-RAR1-dependent or HSP90-SGT1-RAR1-independent, a functional characterization that would either group or distinguish resistance genes from one another. Such a characterization has already been achieved for the known Mla powdery mildew resistance genes in barley (Zhou et al. 2001); and with mutagenized TILLING populations, the development of analogous network-disrupted genetic stocks can readily be achieved in wheat. Indeed, RAR-A1 and RAR-B1 mutants have already been identified within our tetraploid TILLING population (Uauy et al. 2009) and are now being combined for the purpose of such studies (Chen and Dubcovsky, unpublished). By allowing mutation densities that are significantly higher than those possible in diploid species (Uauy et al. 2009), the polyploid nature of wheat greatly facilitates the development of TILLING populations, making TILLING, along with RNAi transgenic strategies, an ideal tool for strategically imposing perturbations to interactome network nodes.
Due to the complexity, flexibility, and redundancy of biological interaction networks, it will be no trivial task to identify other critical nodes with the capacity, under targeted mutagenesis, to drive phenotypic changes (Benfey and Mitchell-Olds 2008) and thus provide meaningful bases for the functional characterization of resistance genes. Thorough characterization of the metabolic pathways induced under partial resistance interactions will be necessary to identify such nodes in the network neighborhoods of cloned resistance genes. Ultimately, a characterization of resistance genes in terms of their associated network nodes may serve as a viable conceptual framework for assessing functional relatedness and predicting the long-term durability of resistance gene combinations.
Extending the interactome: Clues from the pathogen
While local networks of host proteins that interact with identified resistance genes can provide a valuable means of classifying genes in terms of mechanism, the full power of a systems biology-oriented research program on the wheat rusts will be realized only once the interactome is extended to the larger pathosystem by including interactions with pathogen proteins. Host defense responses that limit infection often initiate through the recognition by plant surveillance mechanisms of pathogen-associated molecular patterns, pathogen effectors, or modifications induced by the pathogen in the host (Jones and Dangl 2006). Thus the successful biotrophic lifestyle of the rusts likely depends upon the ability of the pathogen to avoid or suppress host defenses via perturbation of critical nodes of the defense response pathways, the very nodes that hold promise as the basis for the functional classification of resistance genes. By using cloned resistance genes and other host genes within their local interactome neighborhoods as bait to screen yeast-two-hybrid libraries of infected susceptible wheat leaves, it is possible to identify molecules within the pathogen secretome that interact with single host proteins. In this way, pathogen effectors can help pinpoint critical nodes in the disease resistance interactome that are promising targets for subsequent perturbation studies and potential classifiers of resistance gene function.
Other recent discoveries and technological advances promise to further accelerate the discovery rate of wheat rust effectors. The previously known method of isolating haustoria by affinity chromatography from infected plant material (Hahn and Mendgen 1997) can now be combined with NGS to obtain a detailed inventory of Puccinia genes expressed in haustoria. Indeed, expressed sequence tags (ESTs) were recently generated from RNA extracted from isolated haustoria of P. striiformis f. sp. tritici (Yin et al. 2009); and we anticipate that NGS efforts will exponentially increase the number of known haustorially-expressed genes. Bioinformatics tools can then be applied to identify in silico the subset of haustorially-expressed genes that are likely, based on conserved motifs, to be secreted (Emanuelsson et al. 2007). In addition, recent studies in oomycetes and fungi suggest that conserved motifs in the N-terminus of effectors are necessary for delivery of pathogen-produced proteins into plant cells (Dou et al. 2008; Kale et al. 2010). These motifs, thought to be required for binding to particular phosphatidyl inositol phosphates, are hypothesized to facilitate host cell penetration via endocytosis and endosome formation (Kale et al. 2010). By screening the subset of putatively secreted haustorial proteins with these additional motifs, the list of candidate effectors can be restricted further. Once potential effectors are identified, high-throughput functional in planta transient expression assays can be performed to assess their activities and their allelic variants in different host genetic backgrounds, as was recently done with 62 Phytopthora infestans RXLR effector clones (Oh et al. 2009) and with effectors of multiple pathovars of Pseudomonas and Ralstonia spp. (Wroblewski et al. 2009).
Finally, with comparative genomics studies within and among virulent and avirulent races already proving useful in identifying potential pathogenicity factors in Fusarium graminearum (Cuomo et al. 2007; Ma et al. 2010) and Ustilago maydis (Kämper et al. 2006), whole-genome sequencing of Puccinia species also holds much promise. Fully sequenced reference genomes of both P. graminis and P. triticina are annotated and publically available (http://www.broadinstitute.org/annotation/genome/puccinia_group/MultiHome.html); and, from our laboratory, an un-annotated draft of the P. striiformis race PST-130 genome will soon be available through GenBank (Cantu et al. submitted). Such resources open the possibility of surveying across races the variation in pathogen molecules that interact with host proteins. Since resistance response pathways which initiate upon recognition of highly conserved regions of pathogen molecules are more likely to contribute to durable (i.e. race non-specific) resistance strategies than those which initiate upon recognition of more variable regions, such a survey offers a potential means of assessing the race-specificity, and therefore the potential durability, of any given resistance gene.
Gene combinations and predicting the durability of deployed resistance
As illustrated by the complementary gene pair Lr27 + Lr31 (Singh and McIntosh 1984a, 1984b), the combined performance of any given pair or set of rust resistance genes is not strictly predictable at the outset. Therefore, if the ultimate objective is to engineer maximally durable resistance via strategic combinations of resistance genes, each conferring some partial degree of race non-specific protection, the functional characterization described above for single resistance genes will need to be extended systematically to isogenic lines carrying factorial combinations of cloned genes. Toward this end, the molecular characterization of resistance gene combinations should be complemented with classical epistatic interaction trials (e.g. see Kloppers and Pretorius 1997) to assess not only the effects of various gene combinations on the resistance response but also their effects on overall host fitness. While long-term durability of resistance may lie in a strategy of stacking resistance genes that are as functionally diverse as possible, studies in other pathosystems indicate that crosstalk among various defense and abiotic stress pathways, even those that are not constitutively expressed, may lead to overall decreases in host fitness (Bostock 2005). We argue that a core objective of future research on rust resistance should be determining the optimal degree of functional relatedness of resistance genes to insure the long-term durability of deployed gene combinations.
In the meantime, the early transfer of validated and well-mapped QTL combinations to breeding programs will provide an invaluable venue for extensively phenotyping genes and gene combinations over a broad range of environments and genetic backgrounds. In this way, breeding programs can provide an efficient means of systematically assessing the breeding values of these sources of resistance while simultaneously generating the data needed for developing a theory of durability based on functional relatedness. Of course, rigorous evaluation of the effect of cloned resistance gene(s) on host phenotype under disease pressure will ultimately require the development and comparison of nearly isogenic lines in multiple genetic backgrounds.
Conclusions
As the catalog of reported QTL for partial resistance to the wheat rusts continues to swell, the need for a practical guiding theory of resistance gene combinability and predicted durability increases. Such a theory, if achievable, is the long-term promise of a systems biology-oriented research program committed to the functional characterization of partial resistance genes. The necessary genomic resources and molecular technologies to facilitate such an approach in the wheat-rust pathosystem are now available, making it possible to clone partial resistance genes and effectors, to determine their interaction networks, and to systematically perturb critical nodes within those networks. By successfully integrating these core research activities with comparative genomics studies of Puccinia spp. and classical phenotypic, genetic, and epistatic interaction data, the functional relatedness of resistance genes and the variability of their associated pathogen effectors may serve as a powerful organizing principle for probing and eventually understanding the phenomenon of durable resistance.
Acknowledgements
This project was supported in part by the Borlaug Global Rust Initiative (BGRI), the Durable Rust Resistance in Wheat (DRRW) Project, Cornell University, and in part by the National Research Initiative Competitive Grant no. 2008-35318-18654 from the USDA National Institute of Food and Agriculture.
Footnotes
The rapidly migrating and evolving TTKS (or Ug99) lineage of Pgt races, first detected in East Africa in 1999, is an international concern due to the virulence of various of its members to stem rust resistance genes Sr31, Sr24, and Sr36 (Pretorius et al. 2000; Jin et al. 2008, 2009).
Since the year 2000, two closely-related and aggressive groups of races of P. striiformis tritici characterized by virulence to Yr8 and Yr9 have rapidly spread around the globe. Being adapted to warmer temperatures, these races have expanded the historical range of the disease in North America and have contributed to rust epidemics in several parts of the world (Chen 2005, 2007; Hovmoller et al. 2008; Markell and Milus 2008; Milus et al. 2009).
References
- Ayliffe MA, Lagudah ES. Molecular genetics of disease resistance in cereals. Ann Bot. 2004;94:765–773. doi: 10.1093/aob/mch207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azevedo C, Sadanandom A, Kitagawa K, Freialdenhoven A, Shirasu K, Schulze-Lefert P. The RAR1 interactor SGT1, an essential component of R gene-triggered disease resistance. Science. 2002;295:2073–2076. doi: 10.1126/science.1067554. [DOI] [PubMed] [Google Scholar]
- Benfey PN, Mitchell-Olds T. From genotype to phenotype: systems biology meets natural variation. Science. 2008;320:495–497. doi: 10.1126/science.1153716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bent AF, Kunkel BN, Dahlbeck D, Brown KL, Schmidt R, Giraudat J, Leung J, Staskawicz BJ. RPS2 of Arabidopsis thaliana: a leucine-rich repeat class of plant disease resistance genes. Science. 1994;265:1856–1860. doi: 10.1126/science.8091210. [DOI] [PubMed] [Google Scholar]
- Bent AF, Mackey D. Elicitors, effectors, and R genes: the new paradigm and a lifetime supply of questions. Annu Rev Phytopathol. 2007;45:399–436. doi: 10.1146/annurev.phyto.45.062806.094427. [DOI] [PubMed] [Google Scholar]
- Bostock RM. Signal crosstalk and induced resistance: straddling the line between cost and benefit. Annu Rev Phytopathol. 2005;43:545–580. doi: 10.1146/annurev.phyto.41.052002.095505. [DOI] [PubMed] [Google Scholar]
- Brueggeman R, Rostoks N, Kudrna D, Kilian A, Han F, Chen J, Druka A, Steffenson B, Kleinhofs A. The barley stem rust-resistance gene Rpg1 is a novel disease-resistance gene with homology to receptor kinases. Proc Natl Acad Sci USA. 2002;99:9328–9333. doi: 10.1073/pnas.142284999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brueggeman R, Druka A, Nirmala J, Cavileer T, Drader T, Rostoks N, Mirlohi A, Bennypaul H, Gill U, Kudrna D, Whitelaw C, Kilian A, Han F, Sun Y, Gill K, Steffenson B, Kleinhofs A. The stem rust resistance gene Rpg5 encodes a protein with nucleotide-binding-site, leucine-rich, and protein kinase domains. Proc Natl Acad Sci USA. 2008;105:14970–14975. doi: 10.1073/pnas.0807270105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldwell RM, Schafer JF, Compton LE, Patterson FL. Tolerance to cereal leaf rusts. Science. 1958;128:714–715. doi: 10.1126/science.128.3326.714. [DOI] [PubMed] [Google Scholar]
- Cantu D, Govindarajulu M, Kozik A, Wang M, Chen X, Michelmore RW, Dubcovsky J. Next generation sequencing provides rapid access to the genes of wheat stripe rust. 2010 doi: 10.1371/journal.pone.0024230. Submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter AH, Chen XM, Garland-Campbell K, Kidwell KK. Identifying QTL for high-temperature adult-plant resistance to stripe rust (Puccinia striiformis f. sp tritici) in the spring wheat (Triticum aestivum L.) cultivar ‘Louise’. Theor Appl Genet. 2009;119:1119–1128. doi: 10.1007/s00122-009-1114-2. [DOI] [PubMed] [Google Scholar]
- Chen XM. Epidemiology and control of stripe rust (Puccinia striiformis f. sp tritici) on wheat. Can J Plant Pathol. 2005;27:314–337. [Google Scholar]
- Chen XM. Challenges and solutions for stripe rust control in the United States. Aust J Agr Res. 2007;58:648–655. [Google Scholar]
- Cheng P, Chen X. Molecular mapping of a gene for resistance to stripe rust in spring wheat cultivar IDO377s. Phytopathology. 2008;98:S38–S38. doi: 10.1007/s00122-010-1302-0. [DOI] [PubMed] [Google Scholar]
- Chhuneja P, Kaur S, Garg T, Ghai M, Prashar M, Bains NS, Goel RK, Keller B, Dhaliwal HS, Singh K. Mapping of adult plant stripe rust resistance genes in diploid A genome wheat species and their transfer to bread wheat. Theor Appl Genet. 2008;116:313–324. doi: 10.1007/s00122-007-0668-0. [DOI] [PubMed] [Google Scholar]
- Cloutier S, McCallum BD, Loutre C, Banks TW, Wicker T, Feuillet C, Keller B, Jordan MC. Leaf rust resistance gene Lr1, isolated from bread wheat (Triticum aestivum L.) is a member of the large psr567 gene family. Plant Mol Biol. 2007;65:93–106. doi: 10.1007/s11103-007-9201-8. [DOI] [PubMed] [Google Scholar]
- Cuomo CA, Guldener U, Xu JR, Trail F, Turgeon BG, et al. The Fusarium graminearum genome reveals a link between localized polymorphism and pathogen specialization. Science. 2007;317:1400–1402. doi: 10.1126/science.1143708. [DOI] [PubMed] [Google Scholar]
- Cusick ME, Klitgord N, Vidal M, Hill DE. Interactome: gateway into systems biology. Hum Mol Genet. 2005;14(2):R171–181. doi: 10.1093/hmg/ddi335. [DOI] [PubMed] [Google Scholar]
- Dedryver F, Paillard S, Mallard S, Robert O, Trottet M, Negre S, Verplancke G, Jahier J. Characterization of genetic components involved in durable resistance to stripe rust in the bread wheat ‘Renan’. Phytopathology. 2009;99:968–973. doi: 10.1094/PHYTO-99-8-0968. [DOI] [PubMed] [Google Scholar]
- Ding XD, Richter T, Chen M, Fujii H, Seo YS, Xie MT, Zheng XW, Kanrar S, Stevenson RA, Dardick C, Li Y, Jiang H, Zhang Y, Yu FH, Bartley LE, Chern M, Bart R, Chen XH, Zhu LH, Farmerie WG, Gribskov M, Zhu JK, Fromm ME, Ronald PC, Song WY. A rice kinase-protein interaction map. Plant Physiol. 2009;149:1478–1492. doi: 10.1104/pp.108.128298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou D, Kale SD, Wang X, Jiang RH, Bruce NA, Arredondo FD, Zhang X, Tyler BM. RXLR-mediated entry of Phytophthora sojae effector Avr1b into soybean cells does not require pathogen-encoded machinery. Plant Cell. 2008;20:1930–1947. doi: 10.1105/tpc.107.056093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O, Brunak S, von Heijne G, Nielsen H. Locating proteins in the cell using TargetP, SignalP and related tools. Nat Protocols. 2007;2:953–971. doi: 10.1038/nprot.2007.131. [DOI] [PubMed] [Google Scholar]
- Eulgem T. Regulation of the Arabidopsis defense transcriptome. Trends Plant Sci. 2005;10:71–78. doi: 10.1016/j.tplants.2004.12.006. [DOI] [PubMed] [Google Scholar]
- Fiehn O. Metabolomics - the link between genotypes and phenotypes. Plant Mol Biol. 2002;48:155–171. [PubMed] [Google Scholar]
- Flor HH. Inheritance of pathogenicity in Melampsora lini. Phytopathology. 1942;32:653–659. [Google Scholar]
- Flor HH. The complementary gene systems in flax and flax rust. Adv Genet. 1956;8:29–54. [Google Scholar]
- Fu D, Uauy C, Blechl A, Dubcovsky J. RNA interference for wheat functional gene analysis. Transgenic Res. 2007;16:689–701. doi: 10.1007/s11248-007-9150-7. [DOI] [PubMed] [Google Scholar]
- Fu D, Uauy C, Distelfeld A, Blechl A, Epstein L, Chen X, Sela H, Fahima T, Dubcovsky J. A kinase-START gene confers temperature-dependent resistance to wheat stripe rust. Science. 2009;323:1357–1360. doi: 10.1126/science.1166289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazebrook J. Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu Rev Phytopathol. 2005;43:205–227. doi: 10.1146/annurev.phyto.43.040204.135923. [DOI] [PubMed] [Google Scholar]
- Guo Q, Zhang ZJ, Xu YB, Li GH, Feng J, Zhou Y. Quantitative trait loci for high-temperature adult-plant and slow-rusting resistance to Puccinia striiformis f. sp. tritici in wheat cultivars. Phytopathology. 2008;98:803–809. doi: 10.1094/PHYTO-98-7-0803. [DOI] [PubMed] [Google Scholar]
- Hahn M, Mendgen K. Characterization of in planta-induced rust genes isolated from a haustorium-specific cDNA library. Mol Plant Microbe Interact. 1997;10:427–437. doi: 10.1094/MPMI.1997.10.4.427. [DOI] [PubMed] [Google Scholar]
- Hovmoller MS, Yahyaoui AH, Milus EA, Justesen AF. Rapid global spread of two aggressive strains of a wheat rust fungus. Mol Ecol. 2008;17:3818–3826. doi: 10.1111/j.1365-294X.2008.03886.x. [DOI] [PubMed] [Google Scholar]
- Huang L, Brooks S, Li W, Fellers J, Nelson JC, Gill B. Evolution of new disease specificity at a simple resistance locus in a crop-weed complex: reconstitution of the Lr21 gene in wheat. Genetics. 2009;182:595–602. doi: 10.1534/genetics.108.099614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulbert SH, Webb CA, Smith SM, Sun Q. Resistance gene complexes: evolution and utilization. Annu Rev Phytopathol. 2001;39:285–312. doi: 10.1146/annurev.phyto.39.1.285. [DOI] [PubMed] [Google Scholar]
- Ideker T, Galitski T, Hood L. A new approach to decoding life: systems biology. Annu Rev Genomics Hum Genet. 2001;2:343–372. doi: 10.1146/annurev.genom.2.1.343. [DOI] [PubMed] [Google Scholar]
- International Brachypodium Initiative Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature. 2010;463:763–768. doi: 10.1038/nature08747. [DOI] [PubMed] [Google Scholar]
- International Rice Genome Sequencing Project The map-based sequence of the rice genome. Nature. 2005;436:793–800. doi: 10.1038/nature03895. [DOI] [PubMed] [Google Scholar]
- Jin Y, Steffenson BJ, Fetch T. Sources of resistance to pathotype QCC of Puccinia graminis f. sp. tritici in barley. Crop Sci. 1994;34:285–288. [Google Scholar]
- Jin Y, Szabo LJ, Pretorius ZA, Singh RP, Ward R, Fetch T. Detection of virulence to resistance gene Sr24 within race TTKS of Puccinia graminis f. sp tritici. Plant Dis. 2008;92:923–926. doi: 10.1094/PDIS-92-6-0923. [DOI] [PubMed] [Google Scholar]
- Jin Y, Szabo LJ, Rouse MN, Fetch T, Pretorius ZA, Wanyera R, Njau P. Detection of virulence to resistanc gene Sr36 within the TTKS race lineage of Puccinia graminis f. sp. tritici. Plant Dis. 2009;93:367–370. doi: 10.1094/PDIS-93-4-0367. [DOI] [PubMed] [Google Scholar]
- Johnson R. Practical breeding for durable resistance to rust diseases in self-pollinating cereals. Euphytica. 1978;27:529–540. [Google Scholar]
- Johnson R. Durable resistance, definition of, genetic control, and attainment in plant breeding. Phytopathology. 1981;71:567–568. [Google Scholar]
- Johnson R. A critical analysis of durable resistance. Annu Rev Phytopathol. 1984;22:309–330. [Google Scholar]
- Jones JD, Dangl JL. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- Kale SD, Gu B, Capelluto DGS, Dou D, Feldman E, Rumore A, Arredondo FD, Hanlon R, Fudal I, Rouxel T, Lawrence CB, Shan W, Tyler BM. External lipid PI3P mediates entry of eukaryotic pathogen effectors into plant and animal host cells. Cell. 2010;142:284–295. doi: 10.1016/j.cell.2010.06.008. [DOI] [PubMed] [Google Scholar]
- Kämper J, Kahmann R, Bolker M, Ma LJ, Brefort T, et al. Insights from the genome of the biotrophic fungal plant pathogen Ustilago maydis. Nature. 2006;444:97–101. doi: 10.1038/nature05248. [DOI] [PubMed] [Google Scholar]
- Keller B, Feuillet C, Yahiaoui N. Map-based isolation of disease resistance genes from bread wheat: cloning in a supersize genome. Genet Res. 2005;85:93–100. doi: 10.1017/S0016672305007391. [DOI] [PubMed] [Google Scholar]
- Kitano H. Systems biology: a brief overview. Science. 2002;295:1662–1664. doi: 10.1126/science.1069492. [DOI] [PubMed] [Google Scholar]
- Kloppers FJ, Pretorius ZA. Effects of combinations amongst genes Lr13, Lr34 and Lr37 on components of resistance in wheat to leaf rust. Plant Pathology. 1997;46:737–750. [Google Scholar]
- Kolmer JA. Genetics of resistance to wheat leaf rust. Annu Rev Phytopathol. 1996;34:435–455. doi: 10.1146/annurev.phyto.34.1.435. [DOI] [PubMed] [Google Scholar]
- Krattinger SG, Lagudah ES, Spielmeyer W, Singh RP, Huerta Espino J, McFadden H, Bossolini E, Selter LL, Keller B. A putative ABC transporter confers durable resistance to multiple fungal pathogens in wheat. Science. 2009;323:1360–1363. doi: 10.1126/science.1166453. [DOI] [PubMed] [Google Scholar]
- Lu Y, Lan C, Liang S, Zhou X, Liu D, Zhou G, Lu Q, Jing J, Wang M, Xia X, He Z. QTL mapping for adult-plant resistance to stripe rust in Italian common wheat cultivars Libellula and Strampelli. Theor Appl Genet. 2009;119:1349–1359. doi: 10.1007/s00122-009-1139-6. [DOI] [PubMed] [Google Scholar]
- Ma LJ, van der Does HC, Borkovich KA, Coleman JJ, Daboussi MJ, et al. Comparative genomics reveals mobile pathogenicity chromosomes in Fusarium. Nature. 2010;464:367–373. doi: 10.1038/nature08850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markell SG, Milus EA. Emergence of a novel population of Puccinia striiformis f. sp. tritici in Eastern United States. Phytopathology. 2008;98:632–639. doi: 10.1094/PHYTO-98-6-0632. [DOI] [PubMed] [Google Scholar]
- Martens JW, Dunsmore KM, Harder DE. Incidence and virulence of Puccinia graminis in Canada on wheat and barley in 1988. Plant Pathology. 1989;11:424–430. [Google Scholar]
- Martin GB, Brommonschenkel SH, Chunwongse J, Frary A, Ganal MW, Spivey R, Wu T, Earle ED, Tanksley SD. Map-based cloning of a protein kinase gene conferring disease resistance in tomato. Science. 1993;262:1432–1436. doi: 10.1126/science.7902614. [DOI] [PubMed] [Google Scholar]
- Melichar JP, Berry S, Newell C, MacCormack R, Boyd LA. QTL identification and microphenotype characterisation of the developmentally regulated yellow rust resistance in the UK wheat cultivar Guardian. Theor Appl Genet. 2008;117:391–399. doi: 10.1007/s00122-008-0783-6. [DOI] [PubMed] [Google Scholar]
- Metzker ML. Sequencing technologies - the next generation. Nat Rev Genet. 2010;11:31–46. doi: 10.1038/nrg2626. [DOI] [PubMed] [Google Scholar]
- Milus EA, Kristensen K, Hovmoller MS. Evidence for increased aggressiveness in a recent widespread strain of Puccinia striiformis f. sp. tritici causing stripe rust of wheat. Phytopathology. 2009;99:89–94. doi: 10.1094/PHYTO-99-1-0089. [DOI] [PubMed] [Google Scholar]
- Mindrinos M, Katagiri F, Yu GL, Ausubel FM. The A. thaliana disease resistance gene RPS2 encodes a protein containing a nucleotide-binding site and leucine-rich repeats. Cell. 1994;78:1089–1099. doi: 10.1016/0092-8674(94)90282-8. [DOI] [PubMed] [Google Scholar]
- Morsy M, Gouthu S, Orchard S, Thorneycroft D, Harper JF, Mittler R, Cushman JC. Charting plant interactomes: possibilities and challenges. Trends Plant Sci. 2008;13:183–191. doi: 10.1016/j.tplants.2008.01.006. [DOI] [PubMed] [Google Scholar]
- Mustacchi R, Hohmann S, Nielsen J. Yeast systems biology to unravel the network of life. Yeast. 2006;23:227–238. doi: 10.1002/yea.1357. [DOI] [PubMed] [Google Scholar]
- Oh SK, Young C, Lee M, Oliva R, Bozkurt TO, Cano LM, Win J, Bos JI, Liu HY, van Damme M, Morgan W, Choi D, Van der Vossen EA, Vleeshouwers VG, Kamoun S. In planta expression screens of Phytophthora infestans RXLR effectors reveal diverse phenotypes, including activation of the Solanum bulbocastanum disease resistance protein Rpi-blb2. Plant Cell. 2009;21:2928–2947. doi: 10.1105/tpc.109.068247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park CJ, Peng Y, Chen X, Dardick C, Ruan D, Bart R, Canlas PE, Ronald PC. Rice XB15, a protein phosphatase 2C, negatively regulates cell death and Xa21-mediated innate immunity. PLoS Biol. 2008;6:e231. doi: 10.1371/journal.pbio.0060231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paux E, Sourdille P, Salse J, Saintenac C, Choulet F, Leroy P, Korol A, Michalak M, Kianian S, Spielmeyer W, Lagudah E, Somers D, Kilian A, Alaux M, Vautrin S, Berges H, Eversole K, Appels R, Safar J, Simkova H, Dolezel J, Bernard M, Feuillet C. A physical map of the 1-gigabase bread wheat chromosome 3B. Science. 2008;322:101–104. doi: 10.1126/science.1161847. [DOI] [PubMed] [Google Scholar]
- Peng Y, Bartley LE, Chen X, Dardick C, Chern M, Ruan R, Canlas PE, Ronald PC. OsWRKY62 is a negative regulator of basal and Xa21-mediated defense against Xanthomonas oryzae pv. oryzae in rice. Mol Plant. 2008;1:446–458. doi: 10.1093/mp/ssn024. [DOI] [PubMed] [Google Scholar]
- Pretorius ZA, Singh RP, Wagoire WW, Payne TS. Detection of virulence to wheat stem rust resistance gene Sr31 in Puccinia graminis f. sp. tritici in Uganda. Plant Dis. 2000;84:203. doi: 10.1094/PDIS.2000.84.2.203B. [DOI] [PubMed] [Google Scholar]
- Rohila JS, Chen M, Chen S, Chen J, Cerny R, Dardick C, Canlas P, Xu X, Gribskov M, Kanrar S, Zhu JK, Ronald P, Fromm ME. Protein-protein interactions of tandem affinity purification-tagged protein kinases in rice. Plant J. 2006;46:1–13. doi: 10.1111/j.1365-313X.2006.02671.x. [DOI] [PubMed] [Google Scholar]
- Rosewarne GM, Singh RP, Huerta-Espino J, Rebetzke GJ. Quantitative trait loci for slow-rusting resistance in wheat to leaf rust and stripe rust identified with multi-environment analysis. Theor Appl Genet. 2008;116:1027–1034. doi: 10.1007/s00122-008-0736-0. [DOI] [PubMed] [Google Scholar]
- Rubiales D, Niks RE. Characterization of Lr34, a major gene conferring nonhypersensitive resistance to wheat leaf rust. Plant Dis. 1995;79:1208–1212. [Google Scholar]
- Santra DK, Chen XM, Santra M, Campbell KG, Kidwell KK. Identification and mapping QTL for high-temperature adult-plant resistance to stripe rust in winter wheat (Triticum aestivum L.) cultivar ‘Stephens’. Theor Appl Genet. 2008;117:793–802. doi: 10.1007/s00122-008-0820-5. [DOI] [PubMed] [Google Scholar]
- Scofield SR, Huang L, Brandt AS, Gill BS. Development of a virus-induced gene-silencing system for hexaploid wheat and its use in functional analysis of the Lr21-mediated leaf rust resistance pathway. Plant Physiol. 2005;138:2165–2173. doi: 10.1104/pp.105.061861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo NS, Lee SK, Song MY, Suh JP, Hahn TR, Ronald P, Jeon JS. The HSP90-SGT1-RAR1 molecular chaperone complex: a core modulator in plant immunity. J Plant Biol. 2008;51:1–10. [Google Scholar]
- Shirasu K. The HSP90-SGT1 chaperone complex for NLR immune sensors. Annu Rev Plant Biol. 2009;60:139–164. doi: 10.1146/annurev.arplant.59.032607.092906. [DOI] [PubMed] [Google Scholar]
- Singh RP. Association between Gene Lr34 for leaf rust resistance and leaf tip necrosis in wheat. Crop Sci. 1992;32:874–878. [Google Scholar]
- Singh RP, McIntosh RA. Complementary genes for reaction to Puccinia recondita tritici in Triticum aestivum. I: Genetic and linkage studies. Can J Genet Cytol. 1984a;26:723–735. [Google Scholar]
- Singh RP, McIntosh RA. Complementary genes for reaction to Puccinia recondita tritici in Triticum aestivum. II: Cytogenetic studies. Can J Genet Cytol. 1984b;26:736–742. [Google Scholar]
- Singh RP, Huerta-Espino J, Rajaram S. Achieving near-immunity to leaf and stripe rusts in wheat by combining slow rusting resistance genes. Acta Phytopathol et Entomol Hungarica. 2000;35:133–139. [Google Scholar]
- Singh RP, Hodson DP, Jin Y, Huerta-Espino J, Kinyua MG, Wanyera R, Njau P, Ward RW. Current status, likely migration and strategies to mitigate the threat to wheat production from race Ug99 (TTKS) of stem rust pathogen. CAB Reviews: Perspectives in Agriculture, Veterinary Science, Nutrition and Natural Resources. 2006;1:1–13. [Google Scholar]
- Song WY, Wang GL, Chen LL, Kim HS, Pi LY, Holsten T, Gardner J, Wang B, Zhai WX, Zhu LH, Fauquet C, Ronald P. A receptor kinase-like protein encoded by the rice disease resistance gene, Xa21. Science. 1995;270:1804–1806. doi: 10.1126/science.270.5243.1804. [DOI] [PubMed] [Google Scholar]
- Tsunezuka H, Fujiwara M, Kawasaki T, Shimamoto K. Proteome analysis of programmed cell death and defense signaling using the rice lesion mimic mutant cdr2. Mol Plant Microbe Interact. 2005;18:52–59. doi: 10.1094/MPMI-18-0052. [DOI] [PubMed] [Google Scholar]
- Uauy C, Paraiso F, Colasuonno P, Tran RK, Tsai H, Berardi S, Comai L, Dubcovsky J. A modified TILLING approach to detect induced mutations in tetraploid and hexaploid wheat. BMC Plant Biol. 2009;9:115–128. doi: 10.1186/1471-2229-9-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Norman JM, Benfey PN. Arabidopsis thaliana as a model organism in systems biology. Wiley Interdiscip Rev Syst Biol Med. 2009;1:372–379. doi: 10.1002/wsbm.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YS, Pi LY, Chen X, Chakrabarty PK, Jiang J, De Leon AL, Liu GZ, Li L, Benny U, Oard J, Ronald PC, Song WY. Rice Xa21 binding protein 3 is a ubiquitin ligase required for full Xa21-mediated disease resistance. Plant Cell. 2006;18:3635–3646. doi: 10.1105/tpc.106.046730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wroblewski T, Caldwell KS, Piskurewicz U, Cavanaugh KA, Xu H, Kozik A, Ochoa O, McHale LK, Lahre K, Jelenska J, Castillo JA, Blumenthal D, Vinatzer BA, Greenberg JT, Michelmore RW. Comparative large-scale analysis of interactions between several crop species and the effector repertoires from multiple pathovars of Pseudomonas and Ralstonia. Plant Physiol. 2009;150:1733–1749. doi: 10.1104/pp.109.140251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin C, Chen X, Wang X, Han Q, Kang Z, Hulbert SH. Generation and analysis of expression sequence tags from haustoria of the wheat stripe rust fungus Puccinia striiformis f. sp. tritici. BMC Genomics. 2009;10:626. doi: 10.1186/1471-2164-10-626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F, Kurth J, Wei F, Elliott C, Vale G, Yahiaoui N, Keller B, Somerville S, Wise R, Schulze-Lefert P. Cell-autonomous expression of barley Mla1 confers race-specific resistance to the powdery mildew fungus via a Rar1-independent signaling pathway. Plant Cell. 2001;13:337–350. doi: 10.1105/tpc.13.2.337. [DOI] [PMC free article] [PubMed] [Google Scholar]