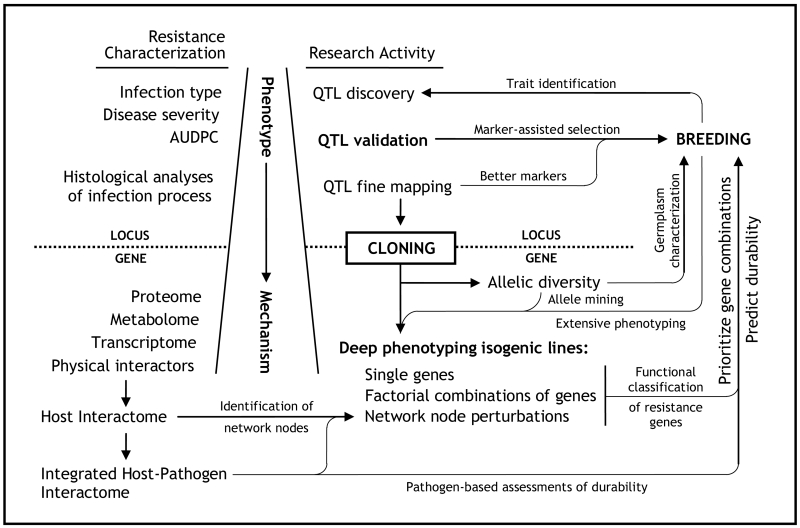
Fig. 1.
Cloning genes underlying broadly effective partial resistance loci is the required point of entry into a research program dedicated to the functional characterization of rust resistance genes in wheat. Cloning permits the rapid assessment of allelic diversity in both research and breeding materials and is thus necessary for the thorough identification, evaluation, and utilization of natural sources of resistance. Cloning also opens the possibility of more extensive and unbiased phenotypic characterizations which take advantage of high-throughput molecular platforms. Once genes are known, their encoded proteins can be placed within interaction networks to which strategic perturbations may be applied, thereby facilitating their functional characterization. Expansion of the interactome to include pathogen proteins offers an additional means of identifying network perturbation targets and assessing the likelihood of true race non-specificity. Integrated with classical phenotypes and rigorous epistatic studies, such information can help prioritize gene combinations for breeding and is likely our best chance to predict and engineer durable resistance. Breeding programs benefit from and are in a position to assist such basic research at all stages, both short- and long-term. (QTL: quantitative trait locus; AUDPC: area under the disease progress curve)