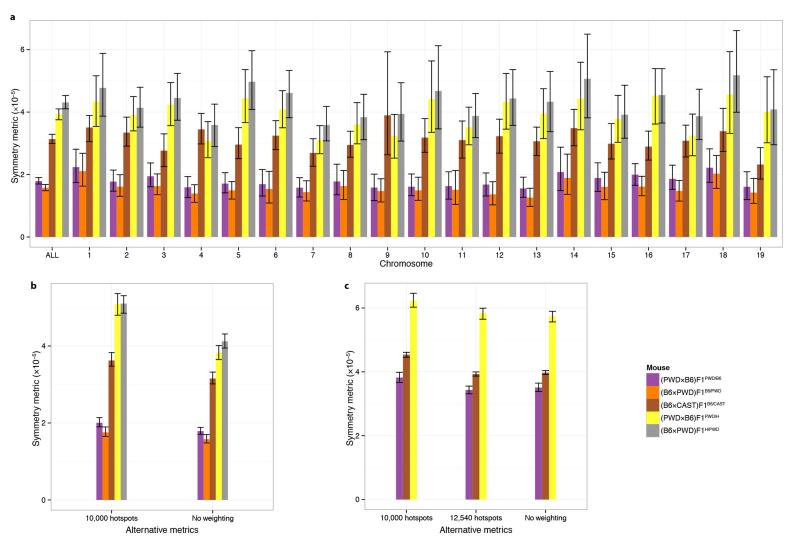
Extended Data Figure 8. Value by chromosome and sensitivity analysis for the symmetry metric.
a, Symmetry metric, as defined in the main text, for each sample (ALL), and for each autosome amongst those samples. Error bars represent bootstrap 95% CIs in all panels. b, Alternative symmetric metrics (to the ones reported in the main text), using only 10,000 hotspots per sample, or without weighting each chromosome specific metric, to compute the average metric genome-wide. Both metrics are computed using the DMC1 maps. c, Alternative symmetric metrics using H3K4me3 maps, similarly to b. The threshold of 12,540 hotspots per sample corresponds to the number of hotspots with ratio estimates in the (PWDxB6)F1PWD/H mouse, which was the lowest amongst the three samples shown here.