Fig. 2.
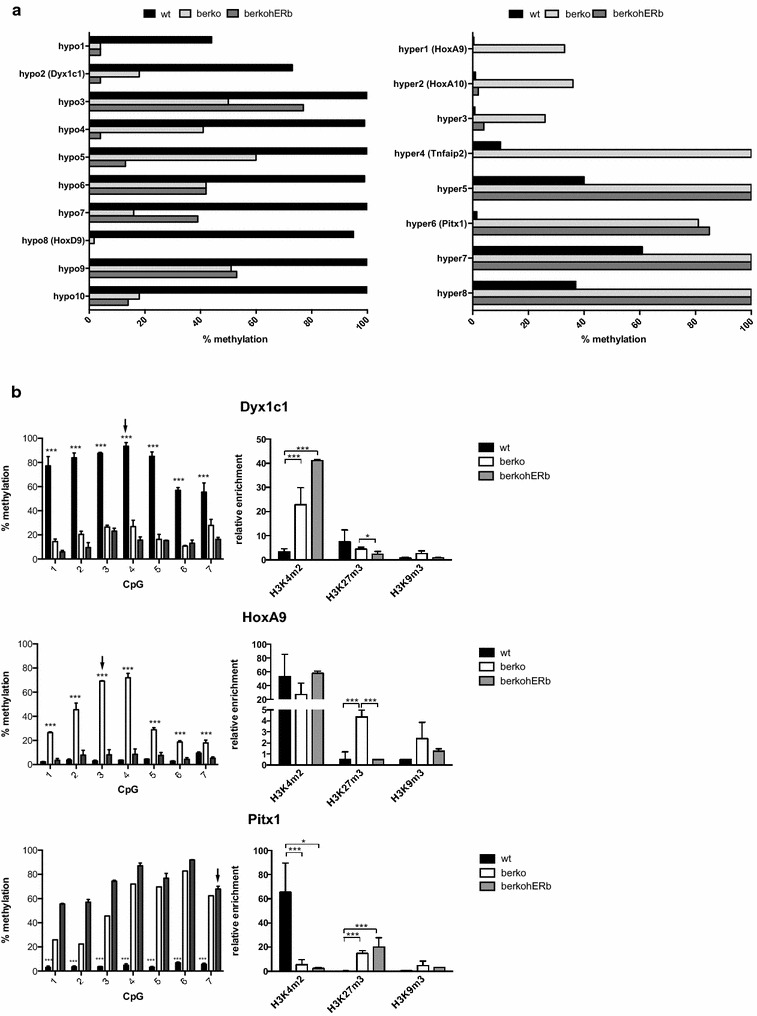
Hyper- but not hypomethylation in βerko MEFs is reversible by re-introduction of ERβ into βerko MEFs. a DNA methylation analysis of ten hypo- and eight hypermethylated positions. DNA methylation was assessed by methylation-specific enzymatic digest followed by qPCR. Positions with gene names in brackets were chosen for further analysis. b DNA methylation (left panel) and histone modifications (right panel) of differentially methylated genes in wt, βerko, and βerkohERβ MEFs. DNA methylation was assessed by pyrosequencing of bisulfite-treated DNA; black arrows mark DMPs identified by RRBS. Triple Asterisk indicates significant differences (p < 0.005) for wt vs. βerko and βerkohERβ (Dyx1c1 and Pitx1) or βerko vs. wt and βerkohERβ (HoxA9). Histone modifications were analysed using ChIP followed by qPCR and normalised to HPRT (H3K4m2 and H3K27m3) or GAPDH promoter (H3K9m3) (means + SD; n ≥ 3)
