Fig. 3.
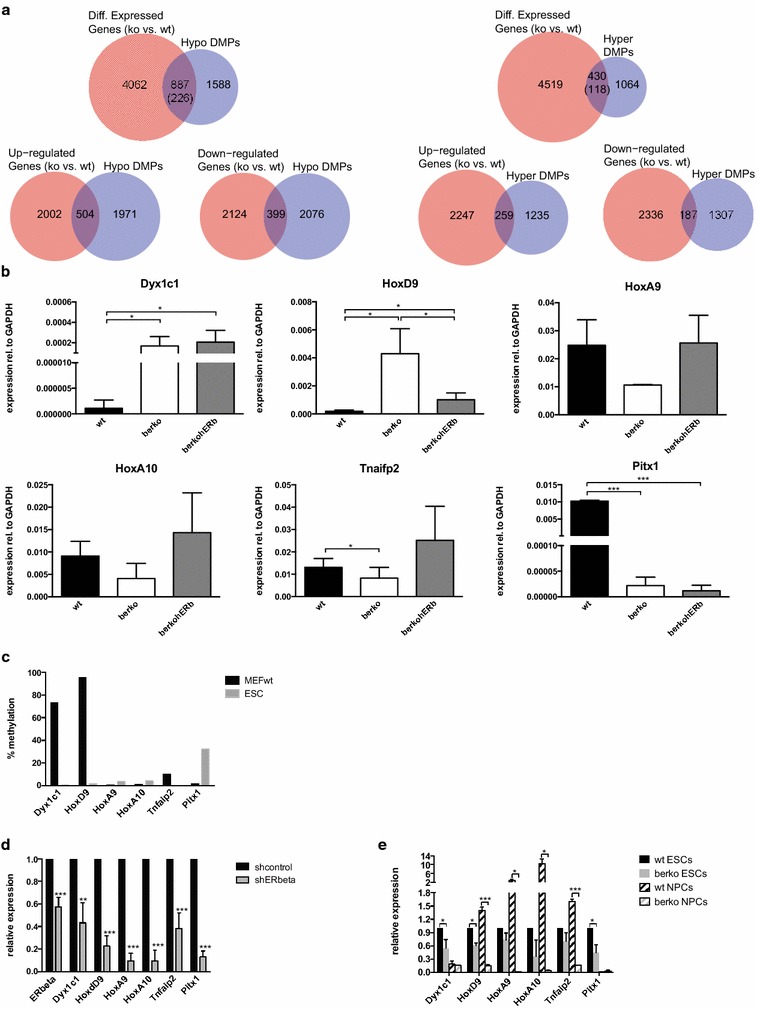
ERβ-dependent transcription of differentially methylated genes in MEFs and ESCs. a Venn diagram visualising overlaps between differentially methylated (identified by RRBS) and differentially expressed (identified by microarray expression analysis) genes in wt and βerko cells. b Gene expression analysis of hypomethylated (Dyx1c1, HoxD9), hypermethylated complementable (HoxA9, HoxA10, and Tnfaip2), and hypermethylated non-complementable genes in wt, βerko, and βerkohERβ MEFs. Gene expression was analysed by RT-qPCR (mean + SD; n ≥ 3). c DNA methylation of differentially methylated genes in wt MEFs and ESCs, assessed by methylation-specific enzymatic digest followed by qPCR. d ERβ-dependent expression of differentially methylated genes in ESCs. Gene expression was assessed by qRT-PCR 4 days after transfection with plasmid encoding for shRNA against ERβ or non-targeting control (means + SD; n ≥ 3). All the genes showed significantly decreased expression compared to shcontrol (** p < 0.01, *** p < 0.005). e ERβ-dependent expression of differentially methylated genes in wt and berko ESCs and NPCs derived thereof. Gene expression was assessed by qRT-PCR (means + SD; n ≥ 3; * p < 0.05, *** p < 0.005)
