Fig. 4.
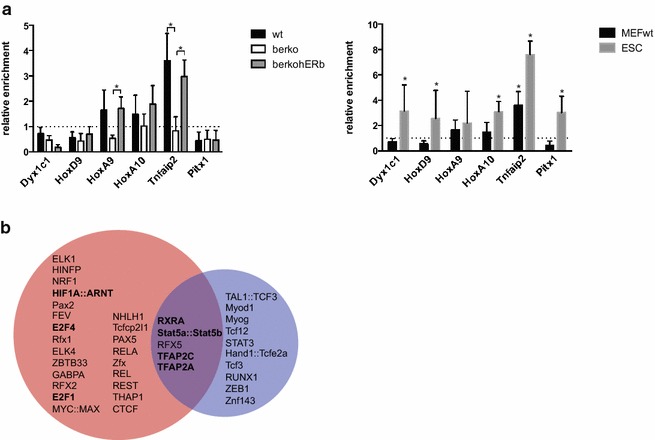
ERβ binds to regions around DMPs. a ERβ recruitment to differentially methylated genes in MEFs (left panel) and ESCs (right panel). HA-tagged ERβ was precipitated and differentially methylated regions were analysed by qRT-PCR. Asterisk indicates significant (p < 0.05) differences between wt/βerkohERβ and βerko MEFs or significant (p < 0.05) enrichment compared to binding at an unrelated, heterochromatic region on chromosome 2 [13] (means + SD; n ≥ 3). b Venn diagram visualising the enriched transcription factor binding motifs around hypo- or hypermethylated DMPs or both. Motifs that have been seen enriched in ERβ-ChIP-seq studies or that are bound by transcription factors known to interact with ERs are depicted in bold
