Fig. 5.
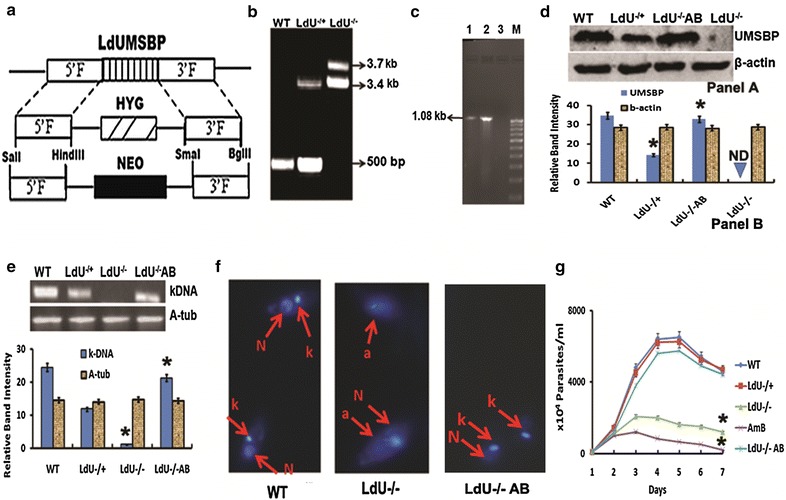
Targeted gene replacement of LdUMSBP alleles to make LdUMSBP depleted parasites and its effect on kDNA and parasite survival. a Schematic representation of the LdUMSBP locus and the plasmid constructs used for gene replacement. b Agarose gel analysis of PCR amplified products of LdUMSBP gene. Lanes WT, LdU−/+ and LdU−/− correspond to PCR with gDNA from WT, LdU−/+ and LdU−/− parasites respectively with external forward and reverse primers which were generated from 42 base pair upstream of LdUMSBP and position 36 downstream of the stop codon of the gene, respectively. The expected size of LdUMSBP, HYG and NEO PCR products are 1.2, 3.4 and 3.7 kb. c The presence of episomal LdUMSBP-pXG-phleo construct in transfected LdU−/+ and LdU−/− parasites were detected by the PCR. Lane 1 and 2 shows the band obtained by PCR using pLp-NEO2 vector specific foreward primer (designed from just upstream of the start code of UMSBP) and LdUMSBP specific reverse primer form LdU−/+ and LdU−/− parasites. Lane 3 is negative control to show the PCR specificity. Lane M is 1 kb marker. Figure shows that the UMSBP deleted parasites contain LdUMSBP ligated in a direct orientation in pXG-phleo vector. d Western blot using anti-LdUMSBP antibody to check the level of protein in indicated parasites. Panel A—Blot image. Panel B—graphical representation of densitometric data e–g Deletion of LdUMSBP induces kDNA loss and decreases parasite survival. e Status of minicircle (kDNA) by semiquantitative RT-PCR of WT, LdU−/+, LdU−/− and LdU−/−AB parasites. Panel A—gel image, Panel B—graphical representation of the densitometry data. f Microscopic analysis of kDNA of WT, LdU−/− and LdU−/−AB parasites respectively. g Effect of UMSBP knockout on promastigotes survival. Single gene deletion has no effect on parasite survival. AmB was used as positive control. Asterisk denotes that the data are significant (p < 0.01)
