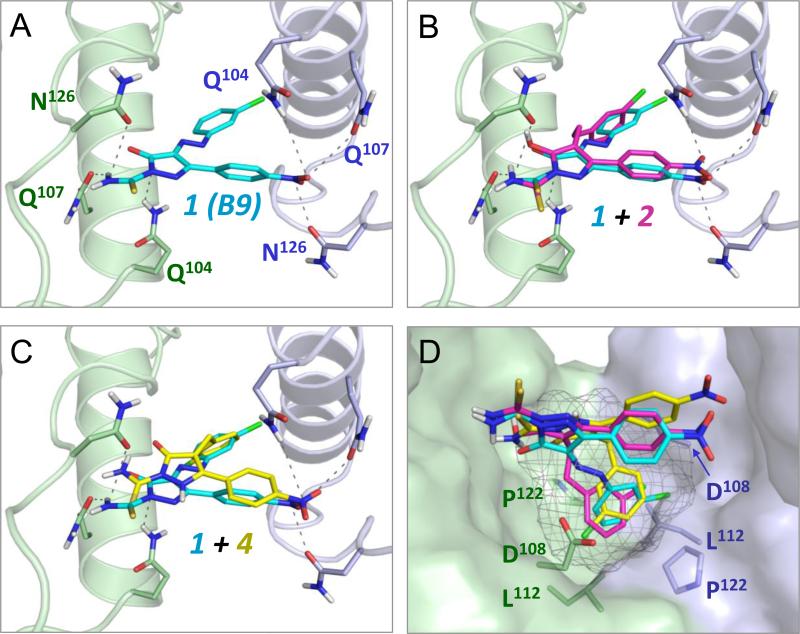
Figure 1.
Computational docking predicts binding of non-azo diphenylpyrazolo inhibitors to the Nef dimer interface. Docking models were generated using the Schrödinger Small Molecule Drug Discovery Suite and the crystal structure of the HIV-1 Nef protein dimer (PDB ID: 1EFN) as described in the Supplementary Information. All four panels show a close-up view of the Nef dimer interface, which is formed by nearly orthogonal interactions of α-helices from each Nef subunit (rendered in blue and green, respectively). A) The parent compound B9 (1, cyan) docks to the Nef dimer interface, forming predicted contacts with conserved Nef residues Gln104, Gln107, and Asn126. This result is virtually identical to a previous docking model produced with Nef and this compound using AutoDock Vina. B and C) Non-azo B9 analogs 2 and 4 dock in a similar pose as B9 at the Nef dimer interface. D) Identification of a hot spot for Nef antagonist binding within the dimer interface (grey mesh). This pocket is defined by the side chains of Nef residues Asp108, Leu112, and Pro122 which accommodate the chlorophenyl group found in all three ligands.