Abstract
The present study aimed at the molecular characterization of pathogenic and non pathogenic F. oxysporum f. sp. lycopersici strains isolated from tomato. The causal agent isolated from symptomatic plants and soil samples was identified based on morphological and molecular analyses. Pathogenicity testing of 69 strains on five susceptible tomato varieties showed 45% of the strains were highly virulent and 30% were moderately virulent. Molecular analysis based on the fingerprints obtained through ISSR indicated the presence of wide genetic diversity among the strains. Phylogenetic analysis based on ITS sequences showed the presence of at least four evolutionary lineages of the pathogen. The clustering of F. oxysporum with non pathogenic isolates and with the members of other formae speciales indicated polyphyletic origin of F. oxysporum f. sp. lycopersici. Further analysis revealed intraspecies variability and nucleotide insertions or deletions in the ITS region among the strains in the study and the observed variations were found to be clade specific. The high genetic diversity in the pathogen population demands for development of effective resistance breeding programs in tomato. Among the pathogenic strains tested, toxigenic strains harbored the Fum1 gene clearly indicating that the strains infecting tomato crops have the potential to produce Fumonisin.
In India, crops are grown under varied agro-climatic conditions and Fusarium spp., occur regularly causing considerable crop losses and produce several mycotoxins1,2. The genetic variability of the members of genus Fusarium increases the difficulties encountered in the development of resistant host genotypes, as well as in effectively deploying available tolerant cultivars. Fusarium oxysporum is a species complex comprising ubiquitous soil borne plant pathogens with more than 150 host-specific forms or formae speciales3,4.
Tomato (Lycopersicon esculentum Mill.) is one of the world’s most widely cultivated vegetable crops for consumption as fresh fruits and various types of processed products5,6. Low yield of tomato is attributed to its susceptibility to several pathogenic fungi, bacteria, viruses and nematodes which are major constraints to tomato cultivation7. Fusarium wilt caused by the soil borne fungus, Fusarium oxysporum Schlectend.: Fr. f. sp. lycopersici (Sacc.) W.C. Snyder and H.N. Hansen, is one of the most devastating diseases of tomato8. It affects greenhouse and field grown tomatoes in warm vegetable production areas. The disease is characterized by yellowed leaves and wilted plants with minimal or absent crop yield. There may be a 30 to 40% yield loss due to the disease and this may go up to 80% under favorable weather conditions9,10. High Fusarium wilt incidence in tomato of 25–55% has been recorded from various parts of India11,12. The pathogen invades the root epidermis and extends into the vascular tissue. It colonizes the xylem vessels producing mycelium and conidia. The characteristic wilt symptoms appear as a result of severe water stress, mainly due to vessel clogging13. Three physiological races (1, 2, and 3) of the pathogen are distinguished by their specific pathogenicity to tomato cultivars14. Since F. oxysporum f. sp. lycopersici (Fol) is an asexual fungus, genetic exchange occurs via somatic fusion and hetreokaryon formation between vegetative compatible strains15.
Chemical treatments and soil solarisation in the fields usually fail to control the pathogen. Use of resistant cultivars is the most reliable method of disease prevention. The selection of an appropriate cultivar requires a thorough understanding of the form and race of the pathogen emerging in the field16. Genetic diversity and phylogenetic analyses within local populations of the pathogen help in elucidating the emergence and evolutionary relationships between the pathogenic and non-pathogenic strains and also may provide information of the pathogen dispersal from other geographical areas17,18. A detailed understanding of the population structure and diversity within Fol is also crucial for the development of effective disease management strategies19. Determining the level of genetic variability within pathogen populations is important since a high genetic variation indicates rapid evolution in response to the changing environmental conditions and the development of new races overcoming host resistance20,21,22.
A variety of approaches had been employed for the characterization of F. oxysporum f. sp. lycopersici isolates. Elias et al.23 studied the population structure of Fol based on RFLP analysis of the genomic DNA which provided genetic evidence that Vegetative Compatibility Grouping (VCG) was an indicator of evolutionary origin. Based on Vegetative Compatibility, mtDNA RFLP, IGS and isozyme polymorphism, several researchers showed that isolates of Fol might represent two genetically distinct evolutionary lineages24,25,26,27. Hirano and Arie28, compared the endopolygalacturonase (pg1) and exopolygalacturonase (pgx4) gene sequences of Fol and Forl from Japan for their differentiation based on PCR. Phylogenetic analyses of sequences of the elongation factor α (EF-1α) and pgx4 gene were conducted by Lievens et al.29 on strains of Fol. Mishra et al.30 determined diversity among Fol isolates based on RAPD. Phoutthasone et al.31 used virulence and AFLP markers for the genetic differentiation between Fol populations of Thailand.
Mycotoxins are the secondary metabolites produced by certain molds on a wide range of agricultural commodities and are closely related to human and animal food chains32,33. Fumonisins are mycotoxins produced by at least 11 species of the fungus Fusarium, including plant pathogens34,35. Fumonisin produced by Fusarium spp., is one of the important toxins which has become a serious constraint in major food crops during the last two decades36. The wide range and frequent presence of Fumonisin toxin found naturally occurring on crops reveal an increasing need for research on the toxigenic potential of Fusarium spp. affecting plants other than cereals to assess the extent of mycotoxin hazard to man and animals.
In view of the economic importance of Fusarium wilt of tomato, the objectives of this study were to investigate phylogenetic relationships between F. oxysporum f. sp. lycopersici and non pathogenic F. oxysporum strains isolated from different tomato growing regions of South India through molecular markers, gene sequence analysis, virulence analysis on different tomato varieties as well as the detection of toxigenic strains.
Results
Morphological identification
All the isolates displayed morphology typical to Fusarium oxysporum. The mycelia of Fusarium oxysporum isolates appeared delicate, white to pink, often with purple tinge, and were sparse to abundant. The fungus produced three types of spores: macroconidia, microconidia and chlamydospores. Macroconidia, sparse to abundant, are borne on branched conidiophores or on the surface of sporodochia and are thin walled, three- to five-septate, fusoid-subulate and pointed at both ends, have pedicellate base. The macroconidia usually measured 15–37.5 μ × 2.5–4 μ and the three-septate macroconidia were more common. Microconidia were abundant, borne on simple phialides arising laterally, oval-ellipsoid, straight to curved, 2.5–15 μ × 2–3 μ usually nonseptate or single septate. Chlamydospores, both smooth and rough walled, were abundant and formed terminally or on an intercalary basis. They are generally solitary, but occasionally form in pairs or chains. The Fusarium oxysporum isolates exhibited a high level of diversity in terms of culture and morphology.
Species specific PCR assay
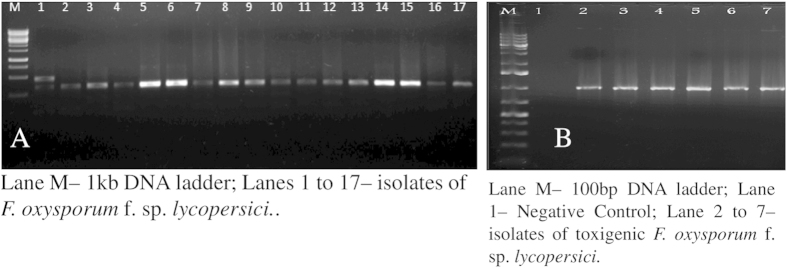
Species specific PCR assay was used for the specific detection of F. oxysporum. PCR with species specific primers amplified a single 340 bp DNA fragment specific to F. oxysporum, thus confirming the species specific identification (Fig. 1A).
Figure 1. Detection of F. oxysporum species and its fumonisin genotypes by PCR assay.
(A) Species-specific PCR assay. (B) Detection of fumonisin positive F. oxysporum species.
Pathogenicity assay
Typical symptoms of wilt disease were first observed 15–20 days after inoculation. In the virulence test, variation in symptoms on aerial parts and within the stem tissues of tomato plants infected with Fol was observed. At early stage, symptoms appeared as yellowing of the lower leaves and in later stages, drooping of the leaves was observed. In severe infection, the pith of the stem was turned brown in colour. In severely infected plants lower leaves dried, ultimately the aerial parts of the tomato plant showed loss of turgidity and drooped down. The results of pathogenicity tests carried out on five cultivars of tomato with 69 isolates of F. oxysporum are presented in Table 1. The F. oxysporum isolates examined had differences in the disease severity on test tomato varieties. Based on the mean disease severity (MDS), the virulence of each isolate was recorded as low (MDS: <25%), moderate (MDS: 25–50%) or high (MDS: >50%). The isolates were thus categorized into 4 groups viz., Highly pathogenic (31 strains), Moderately pathogenic (20 strains), Weakly pathogenic (12 strains) and Non-pathogenic (6 strains) based on the symptomatological variations in the test tomato varieties. The uninoculated tomato seedlings showed no symptoms. Based on the Root-dip inoculation test isolates pathogenic to tomato were identified as F. oxysporum f. sp. lycopersici. All Fol isolates were successfully reisolated from disease-affected plants, thereby completing Koch’s postulates.
Table 1. Strains of Fusarium oxysporum f. sp. lycopersici and non pathogenic strains used for virulence and genetic diversity analysis through the application of ISSR and ITS Sequence analysis.
| Strain No. | Source (Host/Habitat) | Geographic origin | Disease severity observed in tomato cultivars (Disease index 0-100%) |
Virulence grade | GenBankAccession No | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Arka Abha | Arka Alok | Arka Meghali | Arka Saurabh | ArkaVikas | Mean | |||||
| FO38 | Stem | Karnataka | 73.33abc | 80.00abc | 80.00abc | 80.00ab | 80.00a | 78.67 | High | KF914420 |
| FO84 | Stem | Odisha | 53.33cdef | 60.00cdef | 60.00cdef | 60.00bcde | 46.67cde | 56 | High | KF914421 |
| FO81 | Stem | Maharashtra | 13.33hi | 40.00fgh | 40.00fg | 40.00ef | 13.33fg | 29.33 | Moderate | KF914422 |
| FO4 | Stem | Karnataka | 40.00efg | 53.33defg | 53.33def | 40.00ef | 40.00de | 45.33 | Moderate | KF914423 |
| FO3 | Stem | Karnataka | 66.67abcd | 73.33abcd | 73.33abcd | 66.67abcd | 66.67abc | 69.33 | High | KF914424 |
| FO2 | Rhizosphere Soil | Odisha | 13.33hi | 26.67hi | 40.00fg | 26.67fg | 26.67ef | 26.67 | Moderate | KF914425 |
| FO66 | Rhizosphere Soil | Andhra Pradesh | 53.33cdef | 60.00cdef | 46.67efg | 40.00ef | 53.33bcd | 50.67 | High | KF914426 |
| FO93 | Rhizosphere Soil | Andhra Pradesh | 13.33hi | 0j | 26.67gh | 0h | 40.00de | 16 | Low | KF914427 |
| FO56 | Rhizosphere Soil | Tamilnadu | 66.67abcd | 80.00abc | 66.67bcde | 46.67def | 60.00abcd | 64 | High | KF914428 |
| FO52 | Stem | Maharashtra | 13.33hi | 0j | 0i | 40.00ef | 0g | 10.67 | Low | KF914429 |
| FO55 | Rhizosphere Soil | Karnataka | 0i | 13.33ij | 13.33hi | 13.33gh | 0g | 8 | Low | KF914430 |
| FO44 | Rhizosphere Soil | Maharashtra | 0i | 0j | 0i | 0h | 0g | 0 | Non pathogenic | KF914431 |
| FO85 | Rhizosphere Soil | Tamilnadu | 13.33hi | 13.33ij | 13.33hi | 13.33gh | 13.33fg | 13.33 | Low | KF914432 |
| FO14 | Stem | Odisha | 80.00ab | 80.00abc | 80.00abc | 73.33abc | 80.00a | 78.67 | High | KF914433 |
| FO26 | Rhizosphere Soil | Karnataka | 33.33fgh | 46.67efgh | 13.33hi | 40.00ef | 46.67cde | 36 | Moderate | KF914434 |
| FO77 | Rhizosphere Soil | Odisha | 26.67gh | 53.33defg | 53.33def | 40.00ef | 13.33fg | 37.33 | Moderate | KF914435 |
| FO13 | Rhizosphere Soil | Karnataka | 40.00efg | 40.00fgh | 40.00fg | 40.00ef | 40.00de | 40.00 | Moderate | KF914436 |
| FO34 | Stem | Karnataka | 53.33cdef | 60.00cdef | 53.33def | 60.00bcde | 46.67cde | 54.67 | High | KF914437 |
| FO43 | Stem | Karnataka | 60.00bcde | 46.67efgh | 46.67efg | 46.67def | 40.00de | 48 | Moderate | KF914438 |
| FO28 | Rhizosphere Soil | Karnataka | 66.67abcd | 80.00abc | 73.33abcd | 66.67abcd | 73.33ab | 72 | High | KF914439 |
| FO35 | Stem | Karnataka | 0i | 0j | 0i | 0 | 0g | 0 | Non pathogenic | KF914440 |
| FO37 | Stem | Karnataka | 0i | 0j | 0i | 0h | 0g | 0 | Non pathogenic | KF914441 |
| FO60 | Rhizosphere Soil | Odisha | 40.00efg | 40.00fgh | 13.33hi | 40.00ef | 26.67ef | 32 | Moderate | KF914442 |
| FO63 | Rhizosphere Soil | Karnataka | 60.00bcde | 53.33defg | 46.67efg | 60.00bcde | 60.00abcd | 56 | High | KF914443 |
| FO62 | Stem | Karnataka | 0i | 0j | 0i | 0h | 13.33fg | 2.67 | Low | KF914444 |
| FO65 | Stem | Maharashtra | 13.33hi | 40.00fgh | 40.00fg | 40.00ef | 13.33fg | 29.33 | Moderate | KF914445 |
| FO68 | Rhizosphere Soil | Odisha | 73.33abc | 80.00abc | 66.67bcde | 73.33abc | 73.33ab | 73.33 | High | KF914446 |
| FO1 | Stem | Karnataka | 13.33hi | 26.67hi | 13.33hi | 0h | 0g | 10.66 | Low | KF914447 |
| FO9 | Stem | Maharashtra | 0i | 0j | 26.67gh | 13.33gh | 0g | 8 | Low | KF914448 |
| FO22 | Rhizosphere Soil | Karnataka | 0i | 0j | 0i | 0h | 0g | 0 | Non pathogenic | KF914449 |
| FO23 | Rhizosphere Soil | Karnataka | 73.33abc | 80.00abc | 66.67bcde | 60.00bcde | 80.00a | 72 | High | KF914450 |
| FO41 | Rhizosphere Soil | Andhra Pradesh | 80.00ab | 86.67ab | 86.67ab | 86.67a | 80.00a | 84 | High | KF914451 |
| FO30 | Rhizosphere Soil | Maharashtra | 40.00efg | 46.67efgh | 46.67efg | 26.67fg | 40.00de | 40.00 | Moderate | KF914452 |
| FO57 | Rhizosphere Soil | Karnataka | 53.33cdef | 66.67bcde | 60.00cdef | 73.33abc | 60.00abcd | 62.67 | High | KF914453 |
| FO6 | Rhizosphere Soil | Odisha | 13.33hi | 40.00fgh | 40.00fg | 13.33gh | 40.00de | 29.33 | Moderate | KF914454 |
| FO21 | Stem | Karnataka | 73.33abc | 80.00abc | 80.00abc | 66.67abcd | 73.33ab | 74.67 | High | KF914455 |
| FO19 | Root | Karnataka | 73.33abc | 80.00abc | 80.00abc | 66.67abcd | 80.00a | 76 | High | KF914456 |
| FO51 | Stem | Karnataka | 13.33hi | 13.33ij | 0i | 13.33gh | 0g | 8 | Low | KF914457 |
| FO16 | Rhizosphere Soil | Maharashtra | 46.67defg | 46.67efgh | 13.33hi | 40.00ef | 46.67cde | 38.67 | Moderate | KF914458 |
| FO78 | Stem | Karnataka | 80.00ab | 93.33a | 93.33a | 80.00ab | 80.00a | 85.33 | High | KF914459 |
| FO59 | Rhizosphere Soil | Karnataka | 33.33fgh | 40.00fgh | 40.00fg | 0h | 13.33fg | 25.33 | Moderate | KF914460 |
| FO49 | Stem | Karnataka | 80.00ab | 80.00abc | 80.00abc | 80.00ab | 80.00a | 80.00 | High | KF914461 |
| FO54 | Stem | Karnataka | 60.00bcde | 66.67bcde | 66.67bcde | 66.67abcd | 53.33bcd | 62.67 | High | KF914462 |
| FO45 | Rhizosphere Soil | Karnataka | 66.67abcd | 53.33defg | 60.00cdef | 53.33cde | 53.33bcd | 57.33 | High | KF914463 |
| FO79 | Stem | Odisha | 80.00ab | 73.33abcd | 73.33abcd | 80.00ab | 80.00a | 77.33 | High | KF914464 |
| FO69 | Root | Karnataka | 73.33abc | 66.67bcde | 73.33abcd | 80.00ab | 60.00abcd | 70.67 | High | KF914465 |
| FO70 | Rhizosphere Soil | Tamilnadu | 60.00bcde | 60.00cdef | 53.33def | 60.00bcde | 60.00abcd | 58.67 | High | KF914466 |
| FO67 | Stem | Odisha | 80.00ab | 73.33abcd | 80.00abc | 80.00ab | 80.00a | 78.67 | High | KF914467 |
| FO29 | Stem | Karnataka | 0 | 13.33ij | 13.33hi | 0h | 0g | 5.33 | Low | KF914468 |
| FO72 | Stem | Karnataka | 46.67defg | 40.00fgh | 26.67gh | 26.67fg | 53.33bcd | 38.67 | Moderate | KF914469 |
| FO7 | Rhizosphere Soil | Karnataka | 0i | 0j | 0i | 0h | 0g | 0 | Non pathogenic | KF914470 |
| FO10 | Stem | Karnataka | 60.00bcde | 53.33defg | 53.33def | 60.00bcde | 66.67abc | 58.67 | High | KF914471 |
| FO39 | Stem | Maharashtra | 13.33hi | 0j | 13.33hi | 0h | 26.67ef | 10.67 | Low | KF914472 |
| FO31 | Rhizosphere Soil | Karnataka | 53.33cdef | 66.67bcde | 66.67bcde | 53.33cde | 73.33ab | 62.67 | High | KF914473 |
| FO12 | Stem | Karnataka | 13.33hi | 0j | 0i | 0h | 0g | 2.67 | Low | KF914474 |
| FO5 | Stem | Karnataka | 53.33cdef | 60.00cdef | 60.00cdef | 60.00bcde | 53.33bcd | 57.33 | High | KF914475 |
| FO15 | Stem | Karnataka | 80.00ab | 86.67ab | 86.67ab | 86.67a | 80.00a | 84 | High | KF914476 |
| FO105 | Rhizosphere Soil | Andhra Pradesh | 73.33abc | 66.67bcde | 60.00cdef | 66.67abcd | 66.67abc | 66.67 | High | KF914477 |
| FO53 | Stem | Maharashtra | 40.00efg | 40.00fgh | 26.67gh | 26.67fg | 40.00de | 34.67 | Moderate | KF914478 |
| FO42 | Stem | Karnataka | 53.33cdef | 66.67bcde | 53.33def | 60.00bcde | 53.33bcd | 57.33 | High | KF914479 |
| FO20 | Stem | Maharashtra | 0i | 0j | 0i | 0h | 0g | 0 | Non pathogenic | KF914480 |
| FO40 | Stem | Karnataka | 46.67defg | 40.00fgh | 40.00fg | 53.33cde | 60.00abcd | 48 | Moderate | KF914481 |
| FO114 | Rhizosphere Soil | Karnataka | 46.67defg | 40.00fgh | 60.00cdef | 40.00ef | 40.00de | 45.33 | Moderate | KF914482 |
| FO109 | Rhizosphere Soil | Tamilnadu | 46.67defg | 40.00fgh | 53.33def | 46.67def | 60.00abcd | 49.33 | Moderate | KF914483 |
| FO98 | Stem | Karnataka | 60.00bcde | 53.33defg | 60.00cdef | 53.33cde | 60.00abcd | 57.33 | High | KF914484 |
| FO90 | Rhizosphere Soil | Andhra Pradesh | 40.00efg | 0j | 13.33hi | 0h | 40.00de | 18.67 | Low | KF914485 |
| FO83 | Rhizosphere Soil | Maharashtra | 53.33cdef | 60.00cdef | 53.33def | 53.33cde | 60.00abcd | 56 | High | KF914486 |
| FO116 | Rhizosphere Soil | Andhra Pradesh | 46.67defg | 33.33ghi | 60.00cdef | 46.67def | 40.00de | 45.33 | Moderate | KF914487 |
| FO104 | Rhizosphere Soil | Karnataka | 40.00efg | 40.00fgh | 40.00fg | 26.67fg | 40.00de | 37.33 | Moderate | KF914488 |
Disease severity observed in test susceptible tomato cultivars and virulence grade of Fusarium oxysporum strains.
*Disease index for the strains listed, assessed on a 0 to 100% scale. Analysis of variance (ANOVA) was performed and within each column, mean values followed by a common letter do not differ statistically according to DMRT (P < 0.05).
0: healthy plants; 12.5: plants growing regularly with slight vascular discoloration; 25: slight leaf chlorosis and reduced growth, vascular discoloration; 50: chlorosis, 50% growth reduction in respect to the healthy control, vascular discoloration, initial symptoms of wilting; 75: extended vascular discoloration, strong leaf chlorosis, severe growth reduction and wilting symptoms; 100: plants totally wilted and dead.
ISSR fingerprinting
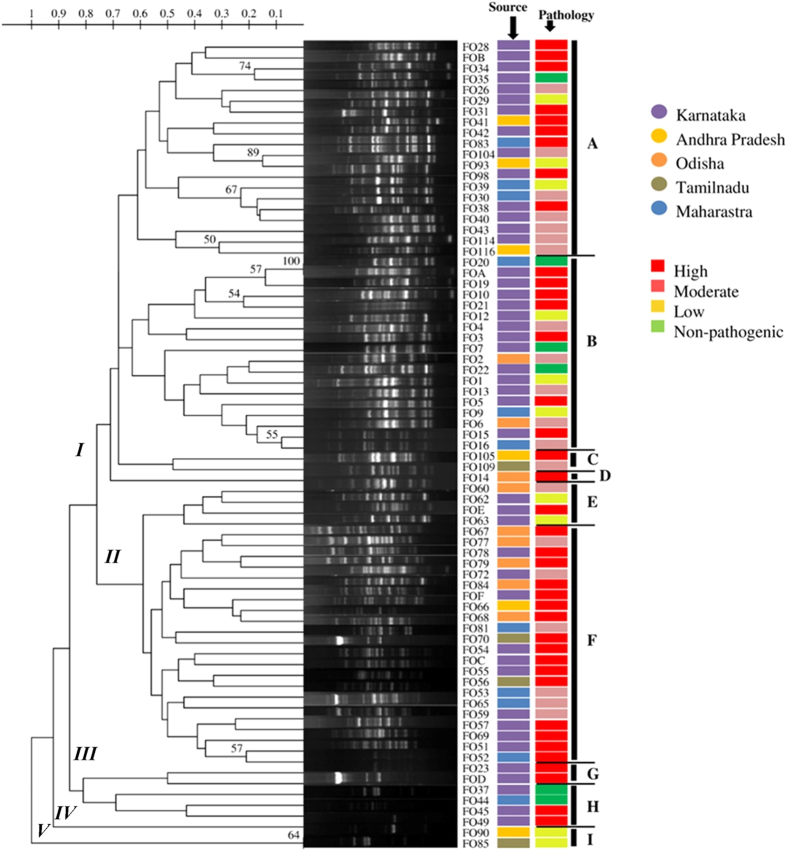
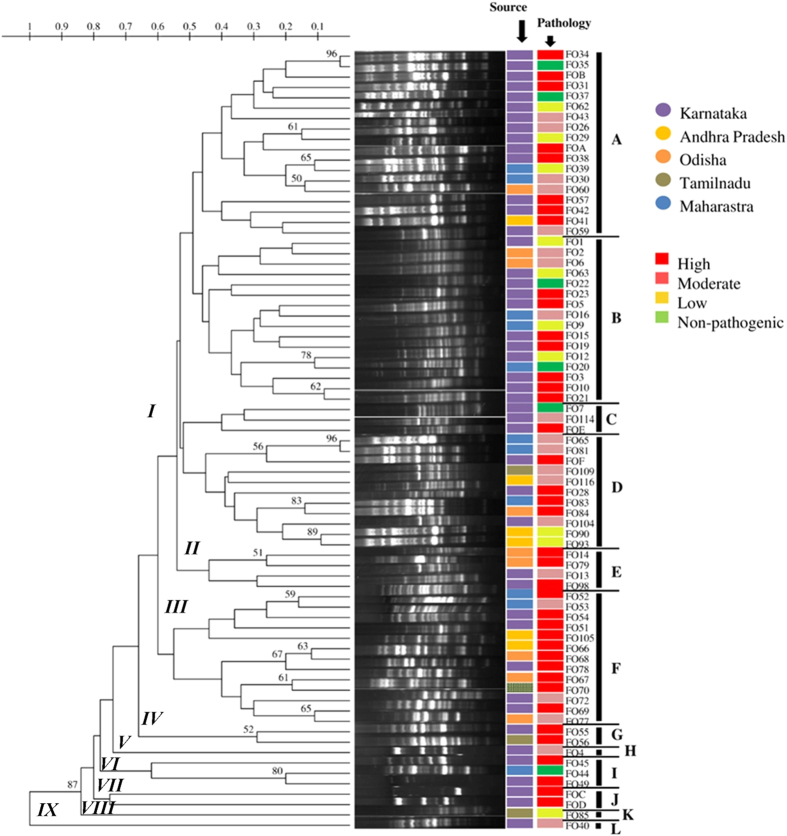
The genetic diversity of F. oxysporum strains was assessed by microsatellite based ISSR fingerprinting technique. A total of 10 ISSR primers were analysed for their capability to produce polymorphic amplicons on a subset of 25 strains including representatives from all the states in the study: Karnataka (39), Andhra Pradesh (6), Odisha (9), Tamilnadu (4) and Maharastra (11). Only 3 primers ((GA)9C, (GA)9T and (CAC)3GC) retained 100% typeability and resulted in robust and reproducible banding patterns for all the strains (Data not shown). Among these primers, two 3’ anchored microsatellites viz., (GA)9C and (GA)9T were utilized in the subsequent studies, the other primer was excluded as it generated smaller number of polymorphic bands allowing insufficient discrimination. The two primers (GA)9C and (GA)9T produced lucid band patterns of a range of approximately 03–26 bands and 02–22 bands, respectively per strain within a range of 100–3000 bp. Using both the primers, all the F. oxysporum strains used in the study were found typeable and UPGMA/Dice dendrograms drawn based on the band patterns showed high diversity among the strains. To determine the discriminatory power of the primers statistically, we estimated Simpson’s (SID) and Shannon’s (H) indices of diversity using Comparing Partitions online tool (Table 2). The calculated SID and H were found equivalent for both the primers at 95% Pearson correlation coefficient level, but on further analysis at lower correlation levels, (GA)9T primer was found more discriminative than (GA)9C primer. Based on the observation of the topology of the dendrograms, major branches of each dendrogram were assigned a roman numeral. The primer (GA)9T resulted in five such major groups (groups I to V) and (GA)9C resulted in nine groups (Groups I to IX) (Figs 2 and 3).
Table 2. Observed subtypes, Simpson’s index of diversity (SID) with jackknife pseudo-values at confidence interval of 95% and entropy (H) at different correlation levels for ISSR typing with two primers used in the study.
| ISSR Primer | Correlation level(%) | # Partitions | SID | Jackknife pseudo-values CI (95%) | H |
|---|---|---|---|---|---|
| (GA)9C | 95 | 72 | 0.999 | (0.998–1.000) | 6.155 |
| 90 | 71 | 0.999 | (0.997–1.000) | 6.122 | |
| 80 | 59 | 0.993 | (0.988–0.998) | 5.765 | |
| 70 | 42 | 0.978 | (0.968–0.988) | 5.114 | |
| 60 | 24 | 0.927 | (0.897–0.956) | 4.025 | |
| (GA)9T | 95 | 73 | 0.999 | (0.998–1.000) | 6.175 |
| 90 | 70 | 0.998 | (0.996–1.000) | 6.095 | |
| 80 | 66 | 0.996 | (0.991–1.000) | 5.959 | |
| 70 | 53 | 0.989 | (0.982–0.995) | 5.559 | |
| 60 | 41 | 0.978 | (0.969–0.987) | 5.096 |
Figure 2. Phylogenetic analysis F. oxysporum f. sp. lycopersici isolates by ISSR analysis with Primer (GA)9T.
Figure 3. Phylogenetic analysis F. oxysporum f. sp. lycopersici isolates by ISSR analysis with Primer (GA)9C.
In the dendrogram generated by banding patterns of (GA)9T-ISSR-PCR, Clade I was the largest and was found to be differentiated into four distinct sub-clades; IA-ID. Majority of the F. oxysporum strains from Karnataka state were grouped in the sub-clades IA and IB (69%) and the rest were distributed across the dendrogram. Clade III had 6 strains and 5 of them were from Karnataka state. All the Odisha strains were clustered in sub-clades IB, ID, IIE and IIF. Strains from AP, TN and MH did not cluster as Karnataka strains and were distributed through the clades. One prominent detail in the dendrogram was one strain each from AP and TN had exactly similar banding pattern and clustered into single clade V and both these isolates were less pathogenic. Another notable difference in banding patterns obtained by (GA)9T-ISSR-PCR from clade V was the presence of an approximately 1300 bp dense band that was absent from the other isolate patterns. The most remarkable feature in the clustering observed in (GA)9T-ISSR-PCR was clade II included only pathogenic strains (Moderate and High) and had at least one representative strain from all the states in the study.
Alternatively, in the dendrogram generated based on banding patterns of (GA)9C-ISSR-PCR, clade I being the largest was differentiated again into four distinct sub-clades, IA-ID. Sub-clades IA, IB and IC contained majorly Karnataka strains and contrastingly only 3 out of 11 strains in ID were from Karnataka. No significant clustering was observed among the strains from different states. One noteworthy pattern observed in the clustering was that clades II, III and IV included only pathogenic strains from all the states and clade ID contained all pathogenic strains except two non-pathogenic strains from AP (FO90 and FO93).
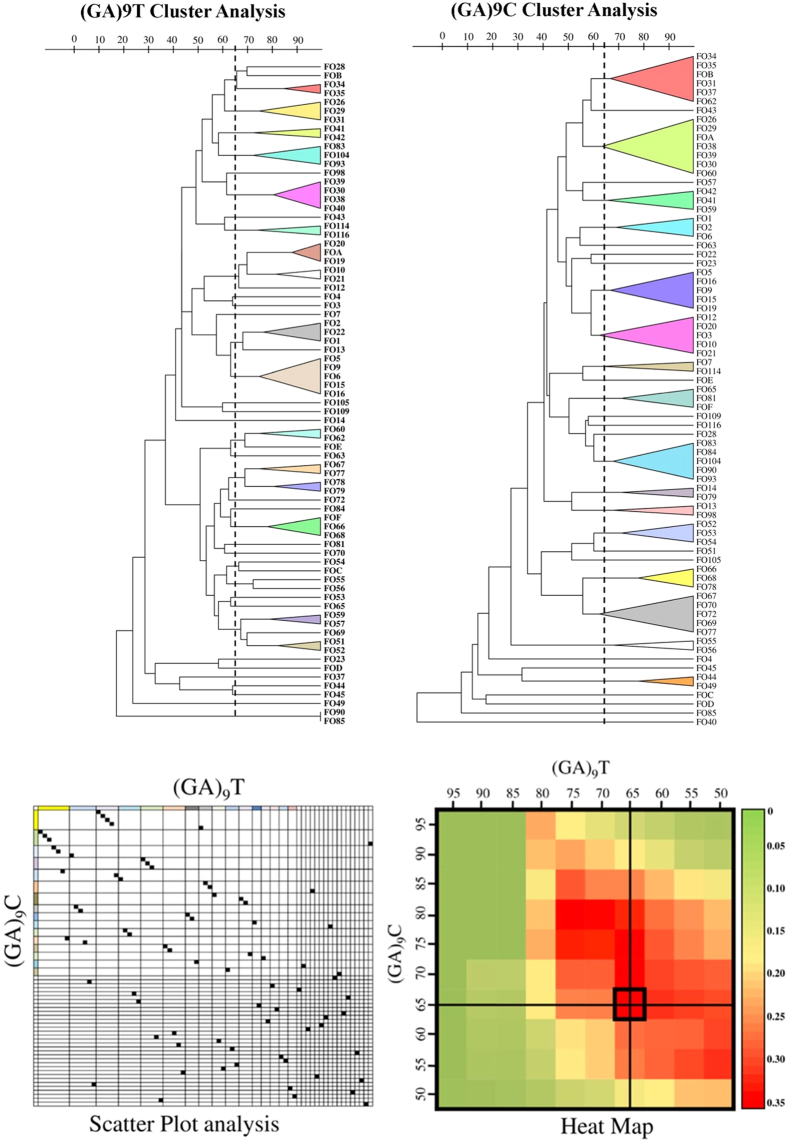
Our next objective was to identify the threshold that better defined the (GA)9T microsatellite partitions in comparison to those defined by (GA)9C. For each possible combination of partitions, the Adjusted Rand value was calculated by varying the threshold cutoffs for each UPGMA/Dice dendrograms of (GA)9C and (GA)9T from 50% to 95%. The maximum coefficient value was determined to be 0.278 at a threshold of 65% for both (GA)9C and (GA)9T dendrograms (Fig. 4). At this cutoff value, 33 clusters were defined by (GA)9C and 48 clusters by (GA)9T. SID values for the partitions found at this threshold level for (GA)9C and (GA)9T were 0.954 and 0.982, respectively. A scatter-plot was also constructed to visually represent the cluster congruence between (GA)9C and (GA)9T clusters and no significant congruence was observed except for four isolates FO5, FO9, FO15 and FO16. For comparing the congruence between type assignments of the (GA)9C and (GA)9T ISSR typing, Wallace coefficients were calculated at 65% cutoff value. Although a bidirectional correspondence, the correlation between the two typing techniques was weak. The Wallace values in the direction of (GA)9C and (GA)9T were 0.452 and 0.226, respectively. Considering this, the probability of two strains that shared a (GA)9T type sharing the same (GA)9C type was only 23% and in vice versa the value was 45%. On a whole, the weak Adjusted Rand and Wallace indices indicated a poor match between the partitions generated by (GA)9C and (GA)9T ISSR typing suggesting that both the microsatellites targeted different loci.
Figure 4. Comparative partition analysis- Diversity and partition congruence between the primers used in this study.
ITS sequence analysis
The ITS region was successfully amplified from DNA from all F. oxysporum strains in the study by the fungal-specific universal primer pairs ITS1–ITS4. The lengths of sequences as determined by capillary electrophoresis ranged from 334 bp to 509 bp. The BLAST analysis of the ITS rDNA sequence data, supported the morphological identification, whereby the closest match (98–100% similarity) in the NCBI GenBank database was found to be with F. oxysporum and Fol. The ITS rDNA sequences of the 69 strains have been deposited in the NCBI GenBank database (GenBank Accession numbers KF914420- KF914488) (Table 1).
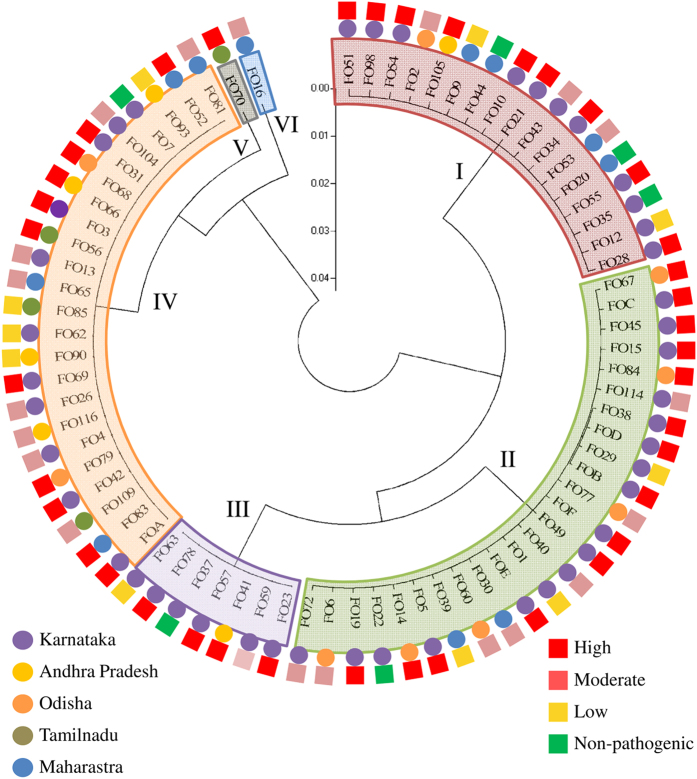
A dendrogram was constructed based on the ITS sequences using ClustalW and Mega5 software by Neighbour-Joining method (Fig. 5). The phylogenetic tree proposed four major clades and two single membered clades with strains distributed across the dendrogram irrespective of their geographic and pathogenic status. Bootstrapping indicated that each branch corresponding to the particular clade was well supported (above 60%). Isolates from Karnataka, Maharashtra, Andhra Pradesh and Odisha were found distributed in the clades I, II and IV. Clade I was found to be heterogenous with isolates from many states and varied pathogenicity. Clade II contained isolates from Karnataka, Odisha and Maharastra states and most of these were pathogenic (high or moderate), except FO22, FO1, FO29 and FO39. Clade III had all the strains isolated from Karnataka except FO41; remarkably this strain was isolated from border region of Andhra Pradesh and Karnataka. Similar to clade I, clade IV was found to be heterogenous. The isolates from Tamilnadu were found distinctly in the clade IV and were absent in clades I, II and III. One pathogenic isolate each from TN and Maharashtra formed the single member clades V and VI, respectively. On a whole, clades I and III typically consisted of pathogenic isolates of Fol isolated in the present study. Interestingly, clade I, III, IV, V and VI uniquely consisted of F. oxysporum f. sp. lycopersici strains. Other formae speciales (reference isolates F. oxysporum f. sp. ciceris, F. oxysporum f. sp. cepae, F. oxysporum f. sp. melonis and F. oxysporum f. sp. phaseoli) were phylogenetically distinct from these clades and clustered only in the clade II along F. oxysporum f. sp. lycopersici. Fifty percent of the non-pathogenic strains were clustered in the clade I and one each were found in clades II, III and IV respectively.
Figure 5. Phylogenetic analysis of F. oxysporum f. sp. lycopersici strains based on rDNA ITS sequences by Neighbor-Joining method.
The tree includes reference strains of F. oxysporum f. sp. lycopersici and strains of other formae speciales.
To appreciate the polymorphism by ITS sequencing, we further studied the sequence analysis, upon which all the strains in the study displayed a general 80% sequence similarity which can be expected due to the conspecific nature of the strains. Further analysis revealed intraspecies variability and nucleotide insertions or deletions in the ITS1 region among the strains. Six variations were identified to be prominent in generation of the diversity observed in the dendrogram, as these variations were found to be clade specific (Supplementary File 1). In general, the variations in the ITS sequences include SNPs, TaqI restriction sites and oligos insertion/deletion termed A to F (Supplimentary File-2).
Detection and Phylogeny of toxigenic F. oxysporum f. sp. lycopersici
A total of 31 highly pathogenic strains were screened to detect and identify fumonisin production. When the primer pair Fum 1 was used, single band having amplicon size of ~790bp was observed for fumonisin producing strains of F. oxysporum (Fig. 1B). Among the 31 selected strains of pathogenic F. oxysporum f. sp. lycopersici tested, 11 isolates were found to be toxigenic harboring the Fum1 gene. The present study reported clearly that some of the F. oxysporum strains infecting tomato have the potential to produce Fumonisin. In the cluster analysis based on ISSR PCR with the primer (GA)9C, the toxigenic strains were found distributed in the clades IA, IIE, IIIF and IVG. Interestingly, in the dendrogram generated with the primer (GA)9C, 60% of the toxigenic strains clustered in the clade IIF and the rest distributed in clades IA and IB. In the phylogenetic tree constructed with NJ based on ITS sequences, 60% of the Fumonisin positive strains clustered in the group IV and the rest were distributed equally in clades I and III.
Discussion
Fusarium wilt of tomato has been reported in India since the late 1960’s but so far there is no comprehensive study on the genetic diversity of the pathogen which occurs virtually in all production areas of the country. In recent years, it has assumed serious proportions in the country due to the failure of recommended management practices made at present and farmers suffer from great economic losses. More recently, the interest in the pathogen has emerged due to increased problems experienced by farmers in tomato production fields. In addition to reporting the presence of Fol, the present study involved determining the genetic diversity within the pathogen population from major tomato growing regions of India.
Field survey revealed high incidence and the widespread prevalence of Fusarium wilt in the major tomato growing regions of India. The Fusarium wilt pathogen Fusarium oxysporum was commonly associated to diseased plants as well as soil samples in the affected fields. The high incidence of Fusarium wilt indicates that this disease is a recurrent problem and that all popular varieties appeared to be susceptible to the disease. Comparison of Fol strains isolated either from infected plants or rhizosphere soils, demonstrated a large diversity in terms of culture and morphology existing among strains of Fol. The strains varied widely in growth, colony characteristics, pigmentation and sporulation. The pathogenicity assay revealed that Fol strains could be categorized into four subgroups viz. non-pathogenic, low virulent, moderate virulent and high virulent strains. The pathogenic strains resulted in the development of the vascular wilt disease in the test tomato cultivars to various levels depending on their virulence. The non-pathogenic strains of F. oxysporum which are efficient colonizers of plant rhizosphere and roots, did not induce any symptoms. The results of the virulence study suggested the existence of a highly variable population of Fol. This study showed that 45% of the strains were highly virulent and 30% were moderately virulent which are distributed in almost all tomato growing areas of Southern India. This poses an important risk to the planting of local tomato varieties due to their susceptibility to the pathogen.
Accurate identification and genetic characterization of pathogens is necessary for appropriate management of plant diseases. The phylogenetic species concept has found relatively recent application in Fusarium systematics and can help to resolve taxonomic difficulties. The ISSR analysis is a robust, PCR-based technique that produces dominant molecular markers by DNA amplification of putative microsatellite regions37. ISSR markers detect a higher level of polymorphism than RAPD markers and have been used extensively in the analysis of fungal population. A high degree of genetic diversity among the Fol strains was observed based on ISSR analysis. Some of the strains formed region specific ISSR patterns and groups in the phylogenetic tree, whereas others from the same region showed distinct patterns and matched with the patterns of the strains of other regions. The ISSR markers clearly indicated the presence of highly variable populations within an area or state of India. However, some of the strains formed region specific ISSR patterns and groups in the phylogenetic tree. The genetic diversity observed among some strains obtained from populations in the same geographic locations were low. This limited variation might probably reflect the nature of the agricultural systems in these areas which greatly rely on the local varieties thus restricting the introduction of new genotypes of the pathogen.
The genetic diversity among the Fol strains of India revealed by ISSR analysis was in concordance with many earlier findings in F. oxysporum from different food crops38. Based on ISSR and other molecular markers, Dubey and Shio39 assessed the genetic diversity of chickpea wilt pathogen which revealed the presence of diverse populations and the occurrence of more than one race of the pathogen in the same locations. Their study indicated the existence of genetically diverse population of F. oxysporum f. sp. ciceris in different states of India. Baysal et al.40 used ISSR and RAPD markers to characterize F. oxysporum f. sp. melongenae isolates. The classification into groups based on ISSR and RAPD fingerprints showed genetic specificity and diversity among the isolates.
The ISSR primers yielded highly polymorphic bands and proved to be authentic and reliable markers for inferring the genetic relationships within the Fol strains. Phylogenetic analysis based on the fingerprints obtained through ISSR analysis indicated the presence of wide genetic diversity among Fol isolates of Southern India. It was evident from our results that ISSR is an efficient technique for the molecular typing of Fol strains and therefore this technique might replace other cumbersome fingerprinting methods for high throughput analysis.
In order to further understand the genetic relationship within the Fol strains, the ITS region was used as a genetic marker to investigate the phylogeny. Phylogenetic analysis based on the ITS sequences helped to reveal the evolutionary relationship within Fol and that with other formae speciales. In the present study, the phylogenetic tree based on the ITS sequence analysis revealed four major groups suggesting four major evolutionary lineages of Fol existing in tomato growing regions of India. Kawabe et al.14 studied a collection Fol isolates from Japan and other countries and reported three evolutionary lineages of Fol detected based on phylogenetic analysis of the ribosomal DNA intergenic spacer (rDNA IGS) region. Each lineage consisted of isolates mainly belonging to a single or closely related vegetative compatibility group (VCG) and a single mating type (MAT)14. Phylogenetic analyses of sequences of the elongation factor α (EF-1α) and exo polygalacturonase (pgx4) gene were conducted by Lievens et al.29 on a worldwide collection of Fol strains. Based on the reconstructed phylogenies, multiple evolutionary lineages were observed in Fol. In addition, mating type analysis showed a mixed distribution of the MAT1-1 and MAT1-2 alleles irrespective of the geographic origin of the isolates. Their study revealed that Fol was composed of at least three independent clonal lineages based on the EF-1α and pgx4 groupings.
Further, the clustering of F. oxysporum f. sp. lycopersici with non pathogenic isolates and with the members of other formae speciales such as F. oxysporum f. sp. ciceris, F. oxysporum f. sp. cepae, F. oxysporum f. sp. melonis and F. oxysporum f. sp. phaseoli was observed in our study. These results suggest that Fol is polyphyletic in origin. O’Donnell et al.41 showed that several formae speciales of F. oxysporum, including Fol were not monophyletic. In other words, isolates of a particular forma specialis might exhibit more close relatedness to some isolates from other formae speciales and non pathogenic strains of F. oxysporum than with the isolates of the same forma specialis26. Based on the present study Fol was found to be polyphyletic suggesting that the pathogenicity towards tomato had evolved several times independently26. These results are in accordance with previous observations of O’Donnell et al. and Cai et al.26,41.
There is a need for rapid, inexpensive and reliable diagnostic protocols for the detection of plant pathogen strains producing different mycotoxins. Although strains of F. oxysporum, do not typically produce fumonisins, there are several reports of fumonisin production by individual strains of this species42,43. PCR approaches based on primer sets targeted to toxin biosynthetic pathway genes have been reported for fumonisinfumonisin producing Fusarium44. In this study we attempted the detection of fumonisin producing Fol using the PCR based approach45. Of the 31 strains isolated from various regions of South India, the primers used detected 11 toxigenic strains of Fol. However, there is a need for further studies focusing on the natural occurrence of fumonisin in diseased tomato plants and fruits and its toxic effects upon consumption.
The ability to distinguish precisely between and within fungal species is crucial in developing a fingerprint database and in determining their emergence and evolution. Genetic differentiation detected within pathogen population can be assumed as environmental factors affecting pathogen properties46. The existence of mixed population for same forma specialis within a region could be due to the possible movement of infested soil on equipment, packing boxes or other items could have introduced the pathogen from one region to the other. Fusarium oxysporum f. sp. lycopersici may be disseminated in seed, transplants, or soil. The tomato crop is produced from seedlings obtained from seedling producers whose seeds are also provided from local and international seed companies38. Evidence from the present study, as well as from literature indicates that microevolutionary events (could be the changes in virulence) occur among strains of Fol belonging to genetically distinct populations. Such changes may occur due to the selection pressure imposed by resistance genes deployed on a large scale in commercial tomato varieties24. Several genes involved in the host defense systems, as well as virulence factors, have been shown to be under positive selection pressure and exhibit a faster accumulation of mutations compared to the genes regulating the basal physiology of the organism47. Adaptive evolutionary shifts of pathogen populations in response to variation in host genotype have been observed in many pathogen-host interactions, e.g., F. oxysporum f. sp. lycopersici on tomato48, F. oxysporum f. sp. vasinfectum on cotton49, F. oxysporum f. sp. dianthi on carnation50. Genetic diversity revealed within same races may be related with the difficulties in controlling the pathogen and the pathogen gaining resistance against the chemicals used in plant protection or may be associated with the exposure of the pathogens to abiotic stress38. Fourie et al.51 suggested that both coevolution with the host and horizontal gene transfer may play important roles in the evolutionary history of the pathogen. Although F. oxysporum is considered to be strictly mitotic, genetic exchange among and within individual lineages might occur more frequently than originally thought52. Genetic recombination could be due to parasexuality, a nonsexual mode of genetic exchange, or heterokaryosis, a process that is initiated by fusion of vegetative hyphae (anastomosis) between individuals with very similar genomes51.
Conclusion
The present study repors the molecular characterization pathogenicity and toxigenicity of F. oxysporum f. sp. lycopersici strains originated from India and their relationship with other formae specilaes within the F. oxysporum complex. The study also indicated that analyzing the genetic variability among Fol strains would be of great importance in plant breeding for disease resistance and can be used by plant breeders. The molecular markers used in the study can be efficiently used for rapid and reliable identification and characterization of Fol population. Our investigation forms the basis for increasing the understanding of underlying genes related to host–pathogen interactions. Such information may help understand virulence dynamics of the pathogen, and facilitate development of efficient disease management strategies using gene-based molecular markers.
Materials and Methods
Fusarium oxysporum strains
Tomato growing areas in 5 states of South India (Karnataka, Andhra Pradesh, Tamilnadu, Maharashtra and Orissa) were surveyed and wilt infected plants and rhizosphere soil samples were collected from 72 different locations. For isolation of Fusarium oxysporum from wilted tomato plant samples, root and stem tissues were washed under running tap water. Plant pieces taken from the lower hypocotyls and upper taproot were surface sterilized in 1% NaOCl (Sodium hypochlorite) solution for 1 to 2 min, rinsed twice in sterile distilled water and dried between sterile filter papers. Pieces of surface disinfected tissues were placed on Potato Dextrose Agar (PDA) plates amended with 50 mg/L tetracycline to suppress the bacterial growth. The plates were incubated at room temperature for 7–10 days for the development of typical white mycelial growth of Fusarium oxysporum and subcultured onto PDA slants53. Isolation from rhizosphere soil samples was done by dilution plate technique. Soil dilutions were plated on PDA and incubated at 28 ± 2 °C for 5 days. Pure cultures were maintained on PDA slants at 4 °C and the lyophilized cultures were stored at −80 °C. The isolates were observed for cultural characters and morphology of the conidia. Colonies exhibiting the taxonomic features of F. oxysporum were identified according to Nelson et al.54. Morphological identification was based on characteristics of the macroconidia, phialides, microconidia, chlamydospores and colony growth traits.
Pathogenicity testing of strains
To determine the formae specials, virulence analysis of the strains was carried out on a set of five tomato cultivars susceptible to Fusarium wilt. A total of 69 strains of F. oxysporum were tested for pathogenicity. Twenty day old healthy seedlings were inoculated by standard root dip method. Seedlings were uprooted carefully preserving the root integrity, shaken to remove the adhering particles and washed gently under tap water. The root apex (about 1 cm) was trimmed with a pair of sterile scissors and submerged for 30 minutes in the conidial suspension of each strain. Seedlings dipped in sterile water served as control. The inoculated seedlings and control were transplanted to minipots (15 cm diameter, surface sterilized with 0.1% mercuric chloride) containing sterilized soil and sand 1:1 ratio. Five seedlings per pot were transplanted. Plants were maintained in a greenhouse where day and night temperatures varied between 25–30 °C. Seedlings were watered daily and fertilized once with NPK (15:15:15). Symptoms started to be visible 15–20 days after artificial inoculation. Disease severity was assessed from 2 weeks of inoculation up to 45 days. The disease index used throughout the experiments ranged from 0 to 100% (0- healthy plant; 25- initial symptoms of leaf chlorosis; 50- severe leaf chlorosis and initial symptoms of wilting; 75-severe wilting symptoms and leaf chlorosis; 100-plant totally wilted, leaves completely necrotic). The discoloration of the vascular tissue was confirmed by slitting the stem24,39. Data were statistically analyzed by using analysis of variance (ANOVA) and Duncan’s test (P < 0.05)55.
DNA Extraction
Fungal strains were grown for 5 days in Potato Dextrose Broth (PDB) at 28 °C under static conditions. Mycelia were harvested and lyophilized. Later 100 mg of the finely powdered mycelium was used for DNA extraction using Hi PurATM Fungal DNA Mini Kit (HiMedia, India), according to the manufacturer’s instructions. The concentration and purity of extracted DNA was determined using NanoDrop-ND-1000 spectrophotometer (Nanodrop Technologies, Wilmington, USA) and the DNA samples were stored at −80 °C for further use.
Species specific PCR assay
Species specific PCR assay was carried out by using the species specific primers as described by Prashant et al.56 (Table 3). The PCR amplification was performed in 20 μL reaction which contained 2 μL of 10× PCR buffer, 2 mM MgCl2, 100 μM dNTPs, 1 unit of Taq polymerase (MBI Fermentas, India) and 20 ηg of template DNA in a Mastercycler thermal cycler (Eppendorf, Hamburg, Germany). The PCR programme consisted of initial denaturation at 94 °C for 4 min followed by 30 cycles of denaturation at 94 °C for 1 min, annealing at 58 °C for 1 min and extension at 72 °C for 2 min, and final extension at 72 °C for 10 min. PCR amplicons were resolved on 1% agarose gel and visualized in a UV transilluminator. A 1 kb ladder was used as a marker. Gels were documented using Gbox (GE health care, Mumbai).
Table 3. List of primers used for identification of F.oxysporum f. sp. lycopersici, fumonisins gene and ITS analysis.
| Primer | Primer Sequence | Amplicon size | Reference |
|---|---|---|---|
| FO1 | 5′-ACATACCACTTGTTGCCTCG-3′ | 340 bp | Prashant et al.2003 |
| FO2 | 5′-CGCCAATCAATTTGAGGAACG-3′ | ||
| Fum1F | 5′-ATTATGGGCATCTTACCTGGAT-3′ | 798 bp | Ramana et al.2011 |
| Fum1R | 5′-ACGCAAGCTCCTGTGACAGA-3′ | ||
| ITS1 | 5′-TCCGTAGGTGAACCTGCGG-3′ | 600 bp | White et al. 1990 |
| ITS4 | 5′-TCCTCCGCTTATTGATATGC-3′ |
Detection of toxigenic F. oxysporum f. sp. lycopersici strains
Primers were used for specific detection of potentially toxigenic F. oxysporum f. sp. lycopersici strains by targeting the metabolic pathway genes specific to fum1 for detection of fumonisin producing strains as reported earlier by Ramana et al.(2011) 44 (Table 3). PCR was carried out in an Eppendorf master cycler gradient (Hamburg, Germany, MastercylcerR pro 384) with a reaction volume of 30 μL. The amplification mixture consisted of template DNA (1.0 μL), 2 mM MgCl2, 1× PCR buffer, 200 μM dNTP mix, 1.0 unit Taq polymerase and primer pairs specific to the targeted gene (fum1) were added at a concentration of 100 nM to the each individual reaction. The PCR was carried out with an initial denaturation at 94 °C for 4 min, followed by 30 cycles of 94 °C for 1 min, 58 °C for 1 min and 72 °C for 1.5 min with a final extension of 72 °C for 8 min. After successful amplification, PCR products were loaded onto 1% agarose gel containing ethidium bromide and visualized under UV.
Genetic diversity among F. oxysporum f. sp. lycopersici strains
Inter Simple Sequence Repeat (ISSR) PCR Analysis
The 69 F. oxysporum f. sp. lycopersici strains subjected to pathogenicity testing were subjected to ISSR analysis. A total of 10 ISSR primers were synthesized from Eurofins, Bangalore, India (Table 4) and used for the amplification of microsatellite loci. To select primers which produce polymorphic and reproducible bands for the characterization of the strains, the 10 ISSR primers with di-or tri nucleotide repeats were screened using DNA samples from representative strains. Among the primers tested, 2 primers (GA)9T and (GA)9C were selected based upon the production of distinct reproducible polymorphic banding patterns. PCR reactions were performed using an Eppendorf master cycler gradient (Hamburg, Germany, MastercylcerR pro 384). PCR was carried out in a 20 μL reaction volume with 2.0 μL of 10× Taq buffer, 0.2 mM dNTPs (2.5 mM each), 2.0 mM MgCl2, 10 pM of each primer, 1.0 U of Taq DNA polymerase and 50 ng template DNA. For each primer-strain combination, amplifications were repeated at least thrice to assure reproducibility. Negative controls using sterile water were included in each experiment. The PCR cycling conditions were carried out with an initial denaturation at 94 °C for 3 min, followed by 35 cycles of 94 °C for 1 min, 45 °C for 1 min and 72 °C for 2 min with a final extension of 72 °C for 10 min. Amplification products were electrophoresed in 1.5% Agarose gel in 0.5× Tris base EDTA buffer at 60 V cm−1.
Table 4. ISSR primers used in the present study to check the genetic diversity among strains of F. oxysporum f. sp. lycopersici.
| Sl No. | PrimerName | Primer Sequence | |
|---|---|---|---|
| 1. | ISSR-1 | CACACACACACAGT | (CA)6GT |
| 2. | ISSR-2 | CTCTCTCTCTCTCTCTTC | (CT)8TC |
| 3. | ISSR-3 | CACCACCACGC | (CAC)3GC |
| 4. | ISSR-4 | CACACACACACACACAA | (CA)8A |
| 5. | ISSR-5 | CACACACACACAAG | (CA)6AG |
| 6. | ISSR-6 | GAGAGAGAGAGAGG | (GA)6GG |
| 7. | ISSR-7 | CTCTCTCTCTCTCTCTGC | (CT)8GC |
| 8. | ISSR-8 | GTGGTGGTGGTGGTG | (GTG)5 |
| 9. | ISSR-9 | GAGAGAGAGAGAGAGAGAC | (GA)9C |
| 10. | ISSR-10 | GAGAGAGAGAGAGAGAGAT | (GA)9T |
Data Analysis
For ISSR analysis, the presence or absence of an allele at a particular locus was scored as 1 and 0 respectively and the pairwise distance among the strains was calculated from the binary matrix using the Jaccard’s coefficient. The resulting distance matrices were used to cluster the strains by the UPGMA (Unweighted Pair Group method using arithmetic means) method.
ITS amplification
The ITS region of the rDNA was amplified using primers ITS1 and ITS4 (White et al. 1990) (Table 3). Amplification reactions were performed in a total volume of 30 μL containing 1× PCR buffer, 200 μM each dNTPs, 1.4 mM MgCl2, 1 μL of each primer (100 nmol), 1.0 U Taq DNA polymerase, and 20 ng of genomic DNA. PCR amplification conditions included an initial denaturation step at 94 °C for 4 min, cycling conditions were 94 °C for 45 s, 56 °C for 45 s, 72 °C for 1 min (30 cycles), followed by a final extension at 72 °C for 5 min. Each set of the experiment included negative controls (without template DNA) to test the presence of contaminating DNA in reagents. The amplicons were checked in 1% agarose gels run parallel to standard DNA molecular weight marker.
ITS Sequence analysis
The amplified ITS rDNA region of selected strains were purified using QIAquick PCR purification kit (QIAGEN, Germany), according to the manufacturer’s instructions. The amplified DNA fragments were then labelled using a Big dye terminator v3.1 cycle sequencing kit and sequenced on a 3730 × l DNA analyzer. The sequences were compared to those in GenBank (http://www.ncbi.nlm.nih.gov/) using Mega BLAST. The BLAST analysis was performed with full length ITS sequences as queries to reveal relationships to published sequences. Highest homology and total score were noted for further analysis. The sequences obtained in the present study were submitted to GenBank. The ITS sequences of F. oxysporum f. sp. lycopersici and F. oxysporum strains from other formae speciales were downloaded from the NCBI GenBank database and were used in the phylogenetic analyses as reference sequences. All the DNA sequences were aligned with the program Clustal W included in BioEdit sequence alignment editor57,58. The resulting multiple-alignment file was used for phylogenetic analyses which were performed using Mega 5.0 with Neighbor-Joining method59.
ITS-RFLP
The ITS PCR products from each of the Fol strains (10 μL) were digested individually with EcoRI restriction enzyme. The reaction mixtures were incubated overnight with the restriction enzyme at 37 °C overnight. The digested products were separated by electrophoresis on 2.0% agarose gel in 0.5 × TBE. The size of the separated fragments was determined by running in parallel a 100 bp DNA ladder (Sigma). Gels were stained with ethidium bromide (0.5 μg ml−1) to visualize the DNA under ultraviolet light and gel images were obtained with a Gel Doc System.
Additional Information
How to cite this article: Nirmaladevi, D. et al. Molecular phylogeny, pathogenicity and toxigenicity of Fusarium oxysporum f. sp. lycopersici. Sci. Rep. 6, 21367; doi: 10.1038/srep21367 (2016).
Supplementary Material
Acknowledgments
The authors are grateful to UGC (University Grants Commission), New Delhi, India for providing the financial support under UGC-Major Research Project F. No. 34-244/2008 (SR).
Footnotes
Author Contributions N.D., V.M., R.K.S., S.K.U., C.N.S., S.C., V.K.G., T.Y.M., C.T.K.M. and N.S.R. designed the experiments; N.D., V.M., S.K.U. and C.N.S. performed experiments; N.D., V.M., C.N.S., V.K.G. and R.K.S. prepared the manuscript; C.N.S., S.C., V.K.G., T.Y.M., C.K.M. and S.R.N. reviewed the manuscript.
References
- Ramana M. V., Nayaka S. C., Balakrishna K., Murali H. S. & Batra H. V. A novel PCR-DNA probe for the detection of fumonisin producing Fusarium species from major food crops grown in southern India. Mycology 3, 167–164 (2012). [Google Scholar]
- Chandra N. S. et al. Prospects of molecular markers in Fusarium species diversity. Appl. Microbiol. Biotechnol. 90, 1625–1639 (2011). [DOI] [PubMed] [Google Scholar]
- Baayen R. P. et al. Gene genealogies and AFLP analyses in the Fusarium oxysporum complex identify monophyletic and nonmonophyletic formae speciales causing wilt and rot disease. Phytopathology 90, 891–900 (2000). [DOI] [PubMed] [Google Scholar]
- Asha B. B., Chandra Nayaka S., Udayashankar A. C., Srinivas C. & Niranjana S. R. Selection of effective bio-antagonistic bacteria for biological control of tomato wilt caused by Fusarium oxysporum. BioScan. 6, 239–244 (2011). [Google Scholar]
- Giovanni C. D. et al. Identification of PCR-based markers (RAPD, AFLP) linked to a novel powdery mildew resistance gene (l-2) in tomato. Plant Sci. 166, 41–48 (2004). [Google Scholar]
- Hariprasad P., Navya H. M., Chandra Nayaka S. & Niranjana S. R. Advantage of PSIRB over PSRB and IRB to improve plant health of tomato. Biol. Cont. 50, 307–316 (2009). [Google Scholar]
- Barone A. & Frusciante L. Marker-assisted selection: current status and future perspectives in crops, livestock, forestry and fish (eds Guimaraes E. et al.) Ch. 9, 151–164 (Agriculture and Consumer Protection Dept., 2007).
- Sudhamoy M., Nitupama M. & Adinpunya M. Salicylic acid induced resistance to Fusarium oxysporum f. sp. lycopersici in tomato. Plant Physiol. Biochem. 47, 642–649 (2009). [DOI] [PubMed] [Google Scholar]
- Kirankumar R., Jagadeesh K. S., Krishnaraj P. U. & Patil M. S. Enhanced growth promotion of Tomato and nutrient uptake by plant growth promoting rhizobacterial isolates in presence of Tobacco Mosaic Virus pathogen. J. Agric. Sci. 21, 309–311 (2008). [Google Scholar]
- Kapoor I. J. Fungi involved in tomato wilt syndrome in Delhi, Maharashtra and Tamilnadu. Indian Phytopathol. 41, 208–213 (1988). [Google Scholar]
- Pandey K. K. & Gupta R.C. Virulence analysis of Fusarium oxysporum f. sp. lycopersici causing tomato wilt in India. J. Mycol. Plant Pathol. 43, 409–413 (2013). [Google Scholar]
- Asha B. B., Chandra Nayaka S., Udayashankar A. C., Srinivas C. & Niranjana S. R. Biological control of Fusarium wilt of tomato. Int. J. Microbiol. Res. 3, 79–84 (2011). [Google Scholar]
- Beckman C. H. The nature of wilt diseases of plants 175–176 (American Phytopathological Society, 1987). [Google Scholar]
- Kawabe M. et al. Three evolutionary lineages of tomato wilt pathogen, Fusarium oxysporum f. sp. lycopersici, based on sequences of IGS, MAT1, and pg1, are each composed of isolates of a single mating type and a single or closely related vegetative compatibility group. J. Gen. Plant Pathol. 71, 263–272 (2005). [Google Scholar]
- Leslie J. F. Fungal vegetative compatibility. Annu. Rev. Phytopathol. 31, 127–150 (1993). [DOI] [PubMed] [Google Scholar]
- Yasushi H. & Tsutomu A. PCR-based differentiation of Fusarium oxysporum f. sp. lycopersici and radicis-lycopersici and races of F. oxysporum f. sp. lycopersici. J Gen. Plant Pathol. 72, 273–283 (2006). [Google Scholar]
- Alves Santos F. M., Benito E. P., Eslava A. P. & Diaz-Minguez J. M. Genetic diversity of Fusarium oxysporum from common bean fields in Spain. Appl. Environ. Microbiol. 65, 3335–3340 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon T. R. & Okamoto D. Variation within and between populations of Fusarium oxysporum based on vegetative compatibility and mitochondrial DNA. Can. J. Bot. 70, 1211–1217 (1992). [Google Scholar]
- Kistler H. C. Evolution of host specificity in Fusarium oxysporum. (eds Summerell B.A. et al.) 70–82 (APS Press, 2001). [Google Scholar]
- McDonald B. A., Miles J., Nelson L. R. & Pettway R. E. Genetic variability in nuclear DNA in field populations of Stagonospora nodorum. Phytopathology 84, 250–255 (1994). [Google Scholar]
- Chandra N. S., Udaya Shankar A. C., Niranjana S. R. & Prakash H. S. Molecular detection and characterization of Fusarium verticillioides in maize (Zea mays. L) grown in Southern India. Ann. Microbiol. 58, 359–367 (2008). [Google Scholar]
- Shanmugam V. & Nandina K. Biological management of vascular wilt of tomato caused by Fusarium oxysporum f. sp. lycospersici by plant growth-promoting rhizobacterial mixture. Biol. Control. 57, 85–93 (2011). [Google Scholar]
- Elias K. S., Zamir D., Lichtmanpleban T. & Katan T. Population-structure of Fusarium oxysporum f. sp lycopersici - restriction fragment length polymorphisms provide genetic evidence that vegetative compatibility group is an indicator of evolutionary origin. Mol. Plant Microbe Interact. 6, 565–572 (1993). [Google Scholar]
- Marlatt M. L., Correll J. C. & Kaufmann P. Two genetically distinct populations of Fusarium oxysporum f. sp. lycopersici race 3 in the United States. Plant Dis. 80, 1336–1342 (1996). [Google Scholar]
- Mes J. J. et al. Biological and molecular characterization of Fusarium oxysporum f. sp. lycopersici divides race 1 isolates into separate virulence groups. Phytopathology 89, 156–160 (1999). [DOI] [PubMed] [Google Scholar]
- Cai G. et al. Origin of race 3 of Fusarium oxysporum f. sp. lycopersici at a single site in California. Phytopathology. 93, 1014–1022 (2003). [DOI] [PubMed] [Google Scholar]
- Attitalla I. H., Jamshid F., Jens L. & Sture B. Rapid molecular method for differentiating two special forms (lycopersici and radicis-lycopersici) of Fusarium oxysporum. Mycol. Res. 108, 787–794 (2004). [DOI] [PubMed] [Google Scholar]
- Hirano Y. & Arie T. PCR-based differentiation of Fusarium oxysporum f. sp. lycopersici and radicis- lycopersici and races of F. oxysporum f. sp. lycopersici. J. Gen. Plant Pathol. 72, 273–283 (2006). [Google Scholar]
- Lievens B. et al. Evolutionary relationships between Fusarium oxysporum f. sp. lycopersici and F. oxysporum f. sp. radicis-lycopersici isolates inferred from mating type, elongation factor-1a and exopolygalacturonase sequences. Mycol. Res. 113, 1181–1191 (2009). [DOI] [PubMed] [Google Scholar]
- Mishra K. K., Ashish K. & Pandey K. K. RAPD based genetic diversity among isolates of Fusarium oxysporum f. sp. lycopersici and their comparative biocontrol. World J. Microbiol. Biotechnol. 26, 1079–1085 (2010). [Google Scholar]
- Phoutthasone S., Jintana U. & Kasem S. Genetic variation of Fusarium oxysporum f. sp. lycopersici isolated from tomatoes in Thailand using pathogenicity and AFLP markers. Afr. J. Microbiol. Res. 6, 5636–5644 (2012). [Google Scholar]
- Ramana M. V., Nayaka S. C., Prakash H. S. & Niranjana S. R. Mycotoxins relevant to bio-warfare and their detection. Toxinology. 1, 295–319 (2014). [Google Scholar]
- Chandra N. S. et al. Laboratory Protocols in Fungal Biology: Current Methods in Fungal Biology (eds Gupta V. K. et al.) Ch. 5, 73–90 (Springer, 2014). [Google Scholar]
- Desjardins A. E. Fusarium Mycotoxins Chemistry, Genetics and Boilogy (APS Press, 2006). [Google Scholar]
- Chandra N. S. et al. Detection and quantification of fumonisins from Fusarium verticillioides in maize grown in southern India. World J. Microbiol. Biotechnol. 26, 71–78 (2010). [Google Scholar]
- Divakara S. T., Santosh M., Aiyaz M., Ramana M. V., Hariprasad P., Chandra Nayaka S. & Niranjana S. R. Molecular identification and characterization of Fusarium spp., associated with sorghum seeds. Journal of Science of Food and Agriculture 94, 1132–1139 (2013). [DOI] [PubMed] [Google Scholar]
- Zietkiewicz E., Rafalski A. & Labuda D. Genomic fingerprinting by simple sequence repeat (SSR) anchored polymerase chain reaction amplification. Genomics 20, 176–183 (1994). [DOI] [PubMed] [Google Scholar]
- Baysal O. et al. Fusarium oxysporum f. sp. lycopersici races and their genetic discrimination by molecular markers in West Mediterranean region of Turkey. Physiol. Mol. Plant Pathol. 74, 68–75 (2009). [Google Scholar]
- Dubey S. C. & Shio R. S. Virulence analysis and oligonucleotide fingerprinting to detect genetic diversity among Indian isolates of Fusarium oxysporum f. sp. ciceris causing chickpea wilt. Mycopathologia 165, 389–406 (2008). [DOI] [PubMed] [Google Scholar]
- Baysal O. et al. Molecular characterization of Fusarium oxysporum f. sp. melongenae by ISSR and RAPD markers on Eggplant. Biochem. Genet. 48, 524–537 (2010). [DOI] [PubMed] [Google Scholar]
- O’Donnell K., Kistler H. C. & Cigelnik P. R. Multiple evolutionary origins of the fungus causing Panama disease of banana: concordant evidence from nuclear and mitochondrial gene genealogies. Proc. Natl. Acad. Sci. USA 95, 2044–2049 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo J. A., Kim J. C. & Lee Y. W. Isolation and characterization of two new type C fumonisins produced by Fusarium oxysporum. J. Nat. Prod. 59, 1003–1005 (1996). [DOI] [PubMed] [Google Scholar]
- Proctor R. H., Busman M., Jeong-Ah S., Lee Y. W. & Plattner R. D. A fumonisin biosynthetic gene cluster in Fusarium oxysporum strain O-1890 and the genetic basis for B versus C fumonisin production. Fungal Genet. Biol. 45, 1016–1026 (2008). [DOI] [PubMed] [Google Scholar]
- Gonzáles-Jaén M. T., Mirete S., Patino B., López-Errasquin. & Vázquez C. Genetic markers for the analysis of variability and for production of specific diagnostic sequences in fumonisin-producing strains of Fusarium verticillioides. Eur. J. Plant Pathol. 110, 525–532 (2004). [Google Scholar]
- Ramana M. V., Balakrishna K., Murali H. S. & Batra H. V. Multiplex PCR-based strategy to detect contamination with mycotoxigenic Fusarium species in rice and finger millet collected from southern India. J. Sci. Food Agric. 91, 1666–1673 (2011). [DOI] [PubMed] [Google Scholar]
- Weller D. M. Biological control of soilborne plant pathogens in the rhizosphere with bacteria. Ann Rev. Phytopathol. 26, 379–407 (1988). [Google Scholar]
- Matute D. R., Quesada-Ocampo L. M., Rauscher J. T. & McEwen J. G. Evidence for positive selection in putative virulence factors within the Paracoccidioides brasiliensis species complex. PLoS Neglected Trop. Dis. 2, 296 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gale L. R., Katan T. & Kistler H. C. The probable center of origin of Fusarium oxysporum f. sp. lycopersici VCG 0033. Plant Dis. 87, 1433–1438 (2003). [DOI] [PubMed] [Google Scholar]
- Wang B., Brubaker C. L., Summerell B. A., Thrall P. H. & Burdon J. J. Local origin of two vegetative compatibility groups of Fusarium oxysporum f. sp. vasinfectum in Australia. Evol. Appl. 3, 505–524 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabanas C. G., Corredor A. V. & Artes E. P. Molecular analysis of Spanish populations of Fusarium oxysporum f. sp. dianthi demonstrates a high genetic diversity and identifies virulence groups in races 1 and 2 of the pathogen. Eur. J. Plant Pathol. 132, 561–576 (2012). [Google Scholar]
- Fourie G., Steenkamp E. T., Gordon T. R. & Viljoen A. Evolutionary relationships among the vegetative compatibility groups of Fusarium oxysporum f. sp. cubense. Appl. Environ. Microbiol. 75, 4770–4781 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor J. W., Jacobson D. J. & Fisher M. C. The evolution of asexual fungi: reproduction, speciation and classification. Ann. Rev. Phytopathol. 37, 197–246 (1999). [DOI] [PubMed] [Google Scholar]
- Norhito Y., Jyuichi S., Mamoru S., Seizo S. & Takashi S. Physiological races and vegetative compatibility groups of butterhead lettuce isolates of Fusarium oxysporum f. sp. lactucae in Japan. J. Gen. Plant Pathol. 70, 308–313 (2004). [Google Scholar]
- Nelson P. E., Toussoun T. A. & Marasas W. F. Fusarium species. An Illustrated manual for identification (Pennsylvania State University press, 1983). [Google Scholar]
- Poli A., Gilardi G., Spadaro D., Gullino M. L. & Garibaldi A. Molecular characterization of Fusarium oxysporum f. sp cichorii pathogenic to chicory (Chicorium intybus). Phytoparasitica 40, 383–391 (2012). [Google Scholar]
- Prashant K. M., Roland T. V. & Alastair C. Development of a PCR-based assay for rapid and reliable identification of pathogenic Fusaria. FEMS Microbiol Lett. 218, 329–332 (2003). [DOI] [PubMed] [Google Scholar]
- Thompson J. D., Higgins D. G. & Gibson T. J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–80 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall T. A. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41, 95–98 (1999). [Google Scholar]
- Tamura K. et al. MEGA5: Molecular Evolutionary Genetics Analysis using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol. Biol. Evol. 28, 2731–2739 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.