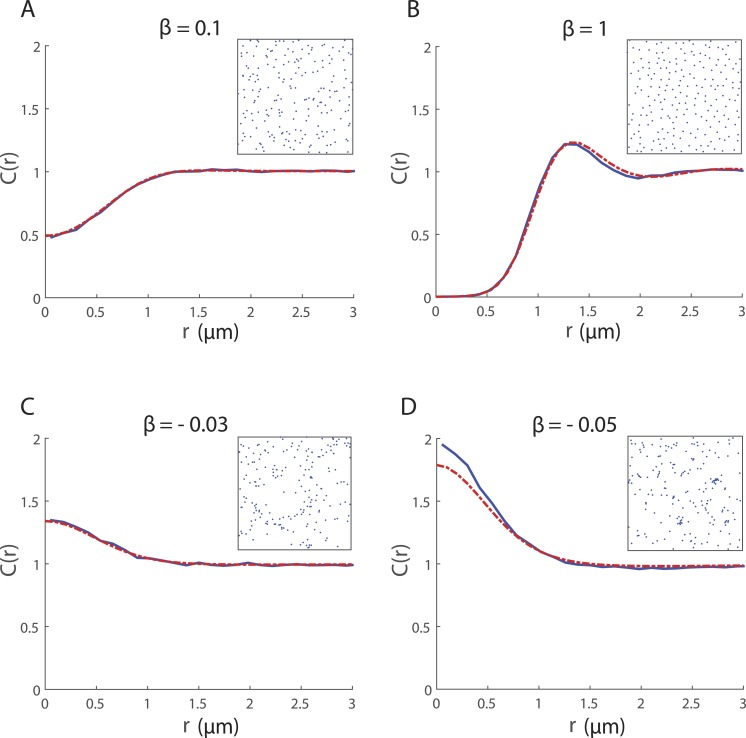
Figure 2. Spatial structure for 200 cells undergoing collective movement with neighbour-dependent directional bias (α = 0h−1) in a 20μ m × 20μ m domain at time t = 25 h.
The PCF CIBM(r) (blue solid line) provides a quantitative measure of the spatial structure in the simulated cell population and is computed (using a bin width δr = 0.12μm) by averaging results from 500 repeated simulations of the IBM. For ease of visualisation, a snapshot of the configuration of cells in a single simulation at t = 25 is shown in the inset. The spatial structure approximated by the spatial moment model (solved using Δ = 0.1μm and ξmax = 4μm) is expressed as a PCF CSM(r) (red dashed line). Parameters are α = 0h−1, σw = σv = 0.5μm, m = 10h−1, λμ = 5μm−1, σμ = 0.05μm; (A) β = 0.1μm; (B) β = 1μm; (C) β = − 0.03μm; (D) β = − 0.05μm.