Figure 2.
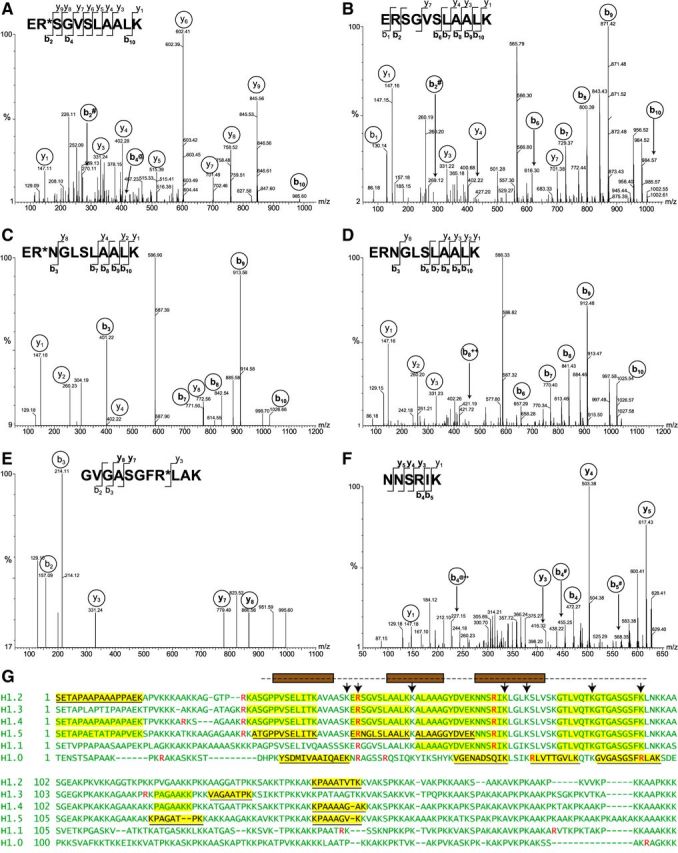
MS/MS analysis of H1 peptides from activated neutrophils A–F) MS/MS spectra of peptides yielding doubly charged precursor ions of H1 histone from Ca2+ ionophore-treated neutrophils. Fragmented ions (b series and y series) are marked on the peptide sequences or spectral peaks. Both peptide ER*SGVSLAALK (A) and ERSGVSLAALK (B) were observed, with a +1 Da shift resulting from deimination of arginine in evidence in b2 and b10 ions. The peptide ERSGVSLAALK is conserved between H1.2, H1.3, and H1.4 subtypes. Both ER*NGLSLAALK (C) and its nondeiminated form ERNGLSLAALK (D) derived from H1.5 were identified. Again, the mass shift of corresponding b ions is consistent with arginine deimination. Arginine-containing peptide GVGASGFR*LAK (E) from H1.0 was also identified, whereas peptide NNSRIK (F), conserved in subtypes H1.1–H1.5, was found only in its nondeiminated form. # indicates -NH3; @ indicates -H2O; ++ indicates a doubly charged ion. G) Summary of results obtained by MS is shown using sequences of 6 human H1 histones. Sequences of somatic linker histone H1 subtypes were aligned to maximize homology. Arginine residues are indicated in red. Peptides highlighted in yellow were identified by mass spectrometry, and H1 subtype-specific peptide sequences are underlined. The globular domain of H1 is indicated by a broken line above the sequences. Brown boxes represent helices that arrange into the H1 winged helix. Residues contributing to DNA binding (50) are indicated with arrows.
