Figure 3.
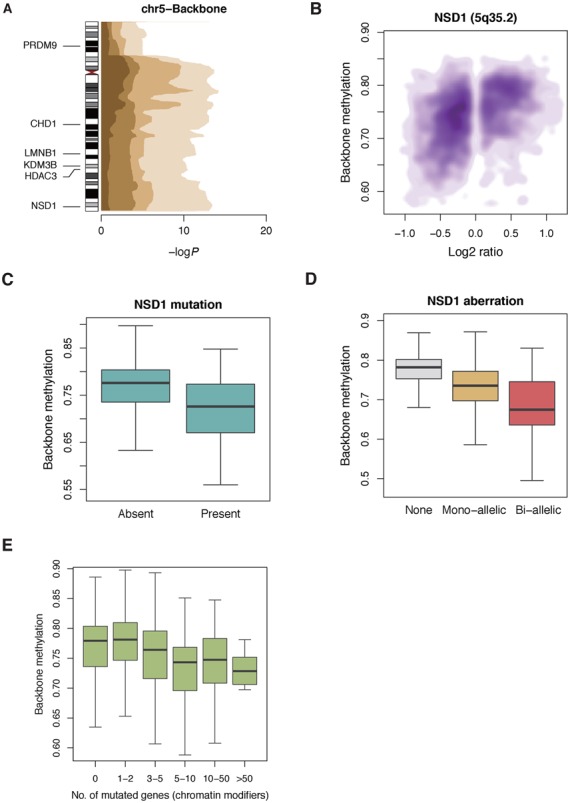
Relation of copy number and somatic mutation with methylation change. (A) By concatenating gene-level associations, copy change at 5q is found to be the most recurrently associated region with backbone demethylation. Color gradients are according to the percentiles of 21 tumor types. Chromatin modifier genes in chromosome 5 are presented. (B) Copy loss of NSD1 at 5q35.2 is significantly correlated with backbone demethylation. All types of tumors analyzed together and copy-neutral tumors were removed from the plot. (C) NSD1-mutated tumors show significant genome-wide backbone demethylation (P = 1.9 × 10−11). (D) Bi-allelic NSD1 aberration by mutation and/or copy loss shows more profound demethylation (P < 2.2 × 10−16). (E) With increasing number of chromatin modifier mutation, more backbone demethylation is apparent (P < 2.2 × 10−16).
