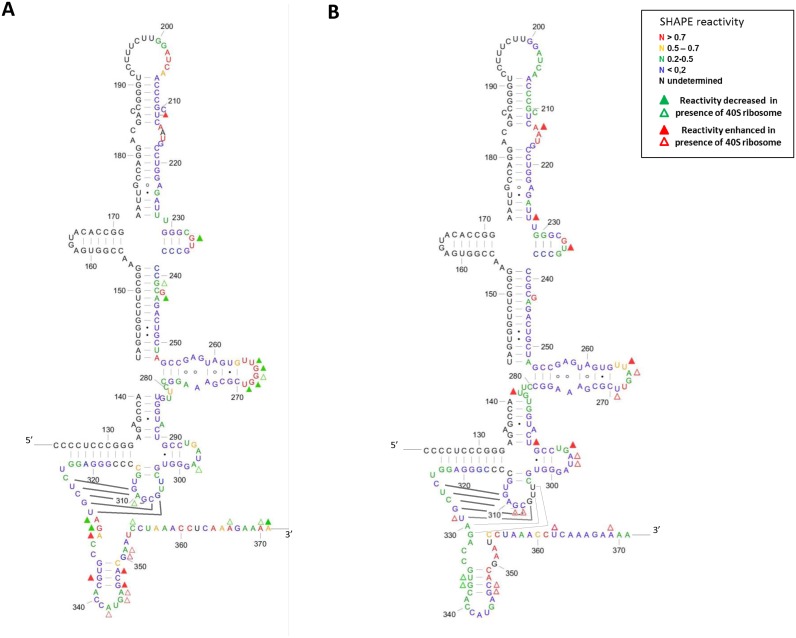
Figure 5.
SHAPE reactivity of the WT (A) and the DM IRES (G266A/G268U) (B) in presence and absence of the 40S ribosomal subunit (400 nM). Relative reactivity calculated with QuSHAPE reported on the secondary structure of the WT and DM IRES. The nucleotides are numbered from the viral transcription initiation start. Only domains III and IV are represented, although the experiment was carried out on the Fluc construct containing domain II and the Fluc coding region (see Materials and Methods). The color code represents the reactivity of each nucleotide as specified in the box. Reactivity values are the mean value from five independent experiments for the WT and five for the DM, they were carried out using two different preparations of both 40S ribosomal subunit and IRESes RNA. Triangles represent footprints observed in presence of the ribosome. The green triangles represent positions for which the reactivity is decreased in presence of 40S (full triangle: t-test < 0.05, ratio r (without 40S/with 40S) >1.5, absolute value of the difference d (|without 40S – with 40S|) >0.1; hollow triangle t < 0.15, r > 2.25, d > 0.15). The red triangles marks positions where the reactivity is enhanced in presence of 40S ribosomal subunit (full triangle t < 0.05, r > 0.6, d > 0.1; hollow triangle t < 0.15, r > 0.4, d > 0.15). The base pair forming the pseudoknot are represented by solid lines, in the mutant the probing data does not exclude that the pseudoknot is extended by 2 base pairs represented as dotted lines. Figures were drawn using XRNA (http://rna.ucsc.edu/rnacenter/xrna/xrna.html).