Figure 5.
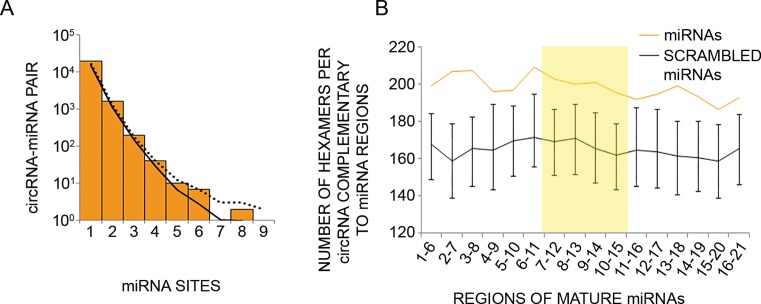
Circular RNAs expressed by human mammary cells show no enrichment for binding sites of the microRNAs found in the same cells. (A) CircRNAs expressed in MCF10A cells were analyzed for the presence of binding sites (7 or 8 nt long) for miRNAs expressed by the same cells (33% uppermost percentile). The number of sites was tallied for each circRNA–miRNA pair, and the distribution of values is plotted. The solid and dotted curves indicate the averaged and 95% upper percentile, respectively, of results when repeating the analysis 1000 times using different permutations of site sequences (see ‘Materials and Methods’ section). (B) Shown are the numbers of hexamers within circRNAs of MCF10A cells that potentially pair to miRNAs expressed in the same cells (orange line), or to scrambled miRNA regions (black line). Error bars represent the distribution of values in 1000 control scrambled miRNA regions. Scrambled miRNA regions preserved mono-nucleotide and GC dinucleotide composition of the original 6 nt window. For each miRNA highly expressed in MCF10A cells (33% upper percentile), starting from the 5′ end, we checked 6-nt-long windows for complementarity to hexamers within the collection of circRNAs expressed in MCF10A cells (N = 1326). The total number of matches was normalized to the number of unique 6 nt windows analyzed. Highlighted is the central region of miRNAs found in a previous study to have statistically significant pairing to coding sequences (55).