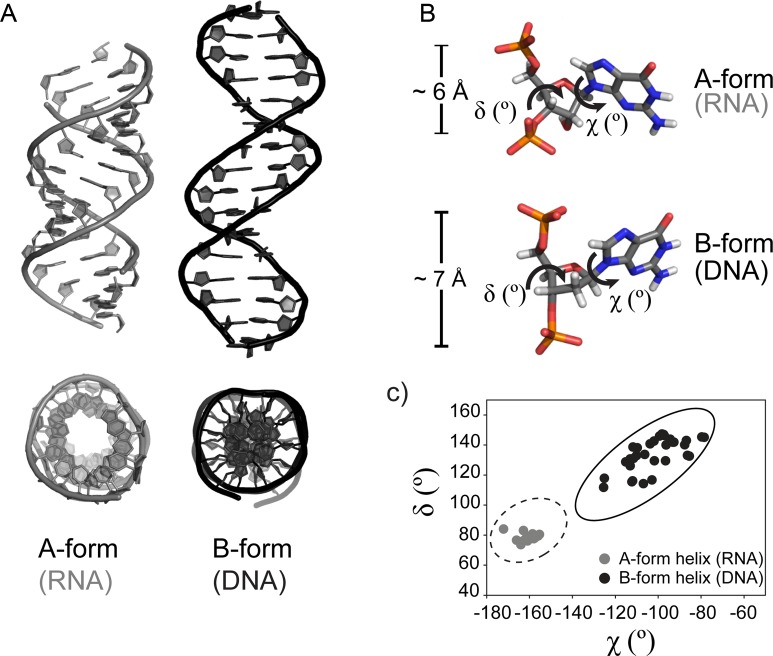
Figure 3.
Geometric parameters of natural A- and B-form helices. (A) Natural genetic polymers (RNA and DNA) adopt different helical geometries. Left: canonical A-form helix (RNA, PDB code 3ND4). Right: canonical B-form helix (DNA, PDB code 3BSE). A view from the top is presented underneath each structure. (B) Differences in sugar puckering and P–P distances between A- and B-form helices. A close-up view of a single nucleotide in a canonical A-helix (top panel, RNA) and a B-helix (bottom panel, DNA) color coded by atom type. The χ and δ torsion angles and the distances between two adjacent phosphates in 5′-3′ direction (PiPi+1) are indicated. (C) (χ,δ) angle covariance matrix for natural A- and B-form helices. Each dot represents a (χ,δ) angle covariance for a single nucleotide from the structures listed above. The values for χ and δ torsion angles of A- and B-form helices cluster in distinct regions of the plot and allow a simple measure of structural diversity. Example values were generated using PYMOL software, based on the PDB code 3ND4 (gray circles, A-form, RNA) and PDB code 3BSE (black circles, B-form, DNA) structures. The ellipses show published value ranges for the (χ,δ) angle covariance matrices of an A-DNA (dashed line) and a B-DNA (solid line) (92).