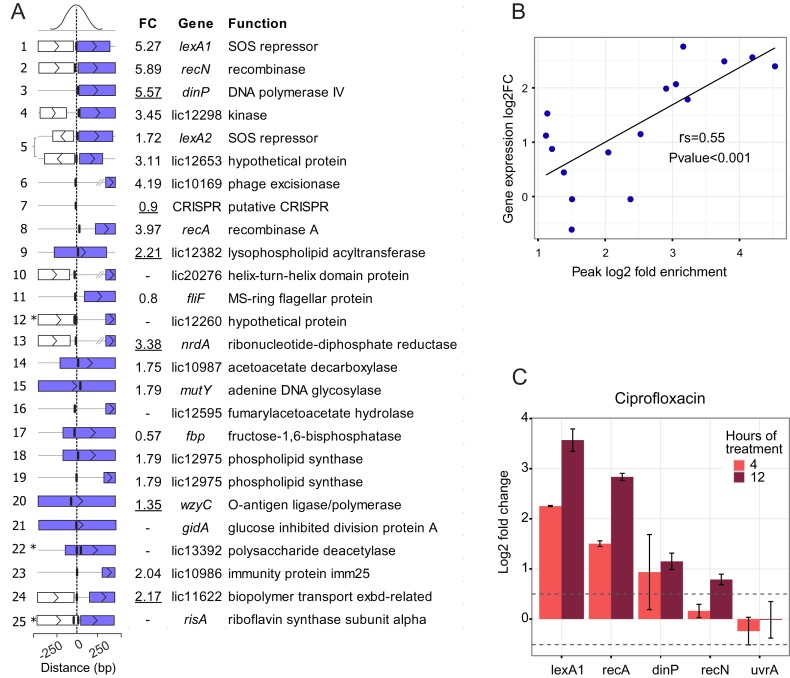
Figure 5.
(A) Genes directly associated with LexA1 binding sites. Peaks are ordered from the most to the least enriched, with the distance from the center of the peak to the next coding region graphically represented. Asterisks indicate peaks containing two closely localized binding sites. Purple rectangles represent the controlled genes, while white ones indicate other ORFs in the vicinity; vertical lines indicate the location of LexA1 binding sites. Differential expression (FC, linear fold change) of genes during the SOS response, as assessed by microarrays or qPCR (underlined values), is shown. (B) Correlation between ChIP-seq peak fold enrichment and log2 fold change of the nearest gene after UV-C damage. Both correlation coefficient (rs) and P-value were computed by Spearman's method. (C) Expression of some core SOS (lexA1, recA, dinP and recN) and not SOS induced (uvrA) genes after 4 or 12h of treatment with 5 μg/ml ciprofloxacin. Values are log2 fold change, calculated by the RT-qPCR 2−ΔΔCt method with six replicates. Error bars represent the standard deviation, and dotted lines comprise fold changes of less than 1.5.