Figure 1.
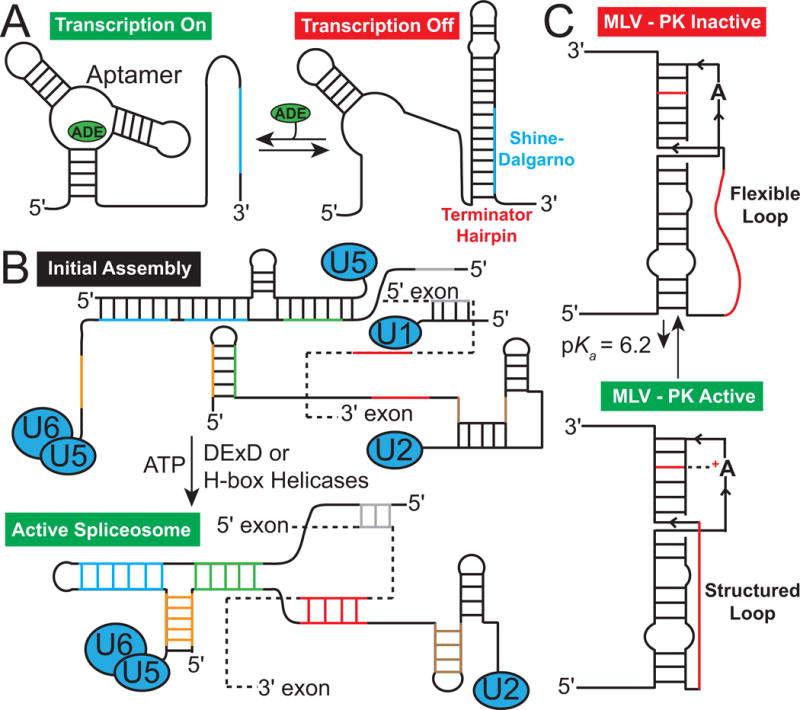
Secondary structural changes in RNA (A) Adenine riboswitch secondary structure with and without the adenine metabolite, which binds to the aptamer domain stabilizing a specific secondary structure turning on transcription. In the absence of metabolite the Shine-Dalgarno sequence (blue) is sequestered turning off transcription (Reining, A. et al., 2013). (B) Secondary structural transitions of mRNA (dashed line) and a spliceosome catalyzed by chaperones (DExD or H-box helicases) that are key for splicing. (C) pH dependent secondary structural equilibrium in the murine leukaemia virus pseudoknot (MLV-PK) mRNA that regulates translational miscoding.
