Figure 5. HOTAIR Overexpression Leads to Androgen-Independent AR Chromatin Targeting.
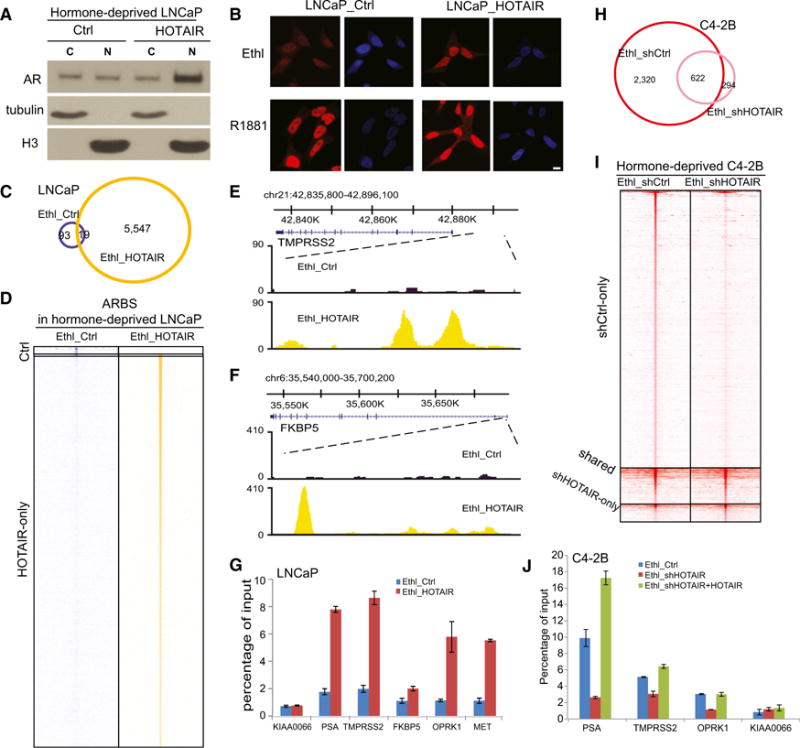
(A) Cytoplasmic and nuclear AR protein levels in hormone-deprived, control and HOTAIR-overexpressing LNCaP cells. Tubulin and H3 were used as cytoplasmic and nuclear protein-loading controls, respectively.
(B) Immunofluorescence staining of AR. LNCaP_ctrl and LNCaP_HOTAIR cells were hormone-starved for 3 days and treated with ethanol or 1 nM R1881 for 24 hr before being subjected to immunostaining using an anti-AR antibody. Scale bar, 10 μm.
(C) Overlap of ARBSs detected in hormone-deprived LNCaP_ctrl and LNCaP_HOTAIR cells is shown.
(D) Heatmap depicts AR ChIP-seq read intensity around (±5 kb) ARBSs shown in (C).
(E and F) Genome browser view of ChIP-seq AR-binding events around AR target genes TMPRSS2 (E) and FKBP5 (F) is shown.
(G) ChIP-qPCR of AR binding on known target genes. AR ChIP was performed in hormone-deprived LNCaP_ctrl and LNCaP_HOTAIR cells. KIAA0066 was the negative control gene. Data shown are mean ± SEM.
(H) Overlap of ARBSs detected in C4-2B cells with control or HOTAIR knockdown is shown.
(I) Heatmap depicts AR ChIP-seq read intensity around (±5 kb) ARBSs shown in (H).
(J) ChIP-qPCR of AR binding on known target genes. AR ChIP was performed in hormone-deprived C4-2B_ctrl, C4-2B shHOTAIR, and C4-2B shHOTAIR + HOTAIR WT cells. KIAA0066 was the negative control gene. Data shown are mean ± SEM.
