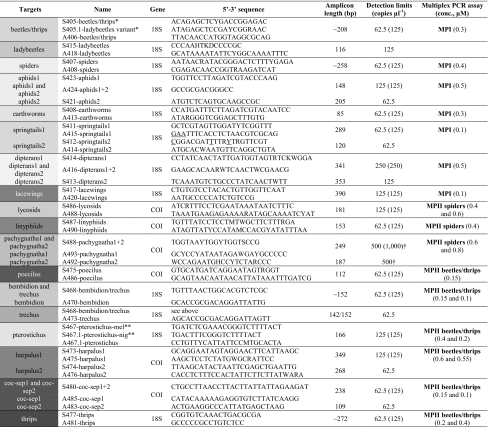
Table 3.
Newly developed primers and the three multiplex PCR assays for assessing trophic interactions of invertebrates in cereal crops. Columns show the primer targets, primer names (S and A denote forward and reverse primers, respectively), targeted gene, primer sequences, expected amplicon lengths, detection limits, and final concentration (conc.) of each primer when used in the multiplex PCR assays (MPI, MPII spiders, and MPII beetles/thrips; if concentrations of forward and reverse primer are different, both are listed). Detection limits refer to the lowest numbers of double-stranded template molecules (copies per µl DNA template) where a detectable amplicon could be generated (i.e. signal strength ≥0.075 RFUs; QIAxcel) in singleplex and optionally multiplex PCR (in parenthesis). The primers marked with * and ** were 1:1 mixes of the two forward primer variants. S411-springtails primer was developed by the authors and published elsewhere (Roubinet et al. 2015); A415 and S412 are slightly modified versions of springtail-primers Col-gen-A246 (Sint et al. 2012) and Col3F (Kuusk and Agusti 2008), respectively (modifications apply to underlined bases). Note that for Trechus amplicon length varies between the two species: T. quadristriatus, 142 bp, T. secalis, 152 bp; the two closely related genera Bembidion and Trechus share the same forward primer (S468)
|
† Due to quality issues of the DNA template for Pachygnatha spp., the sensitivity of the respective primer pairs was additionally tested with highly diluted (1:1000) DNA extracts of Pachygnatha clercki where always very strong signals were produced (>2.4 RFUs)
