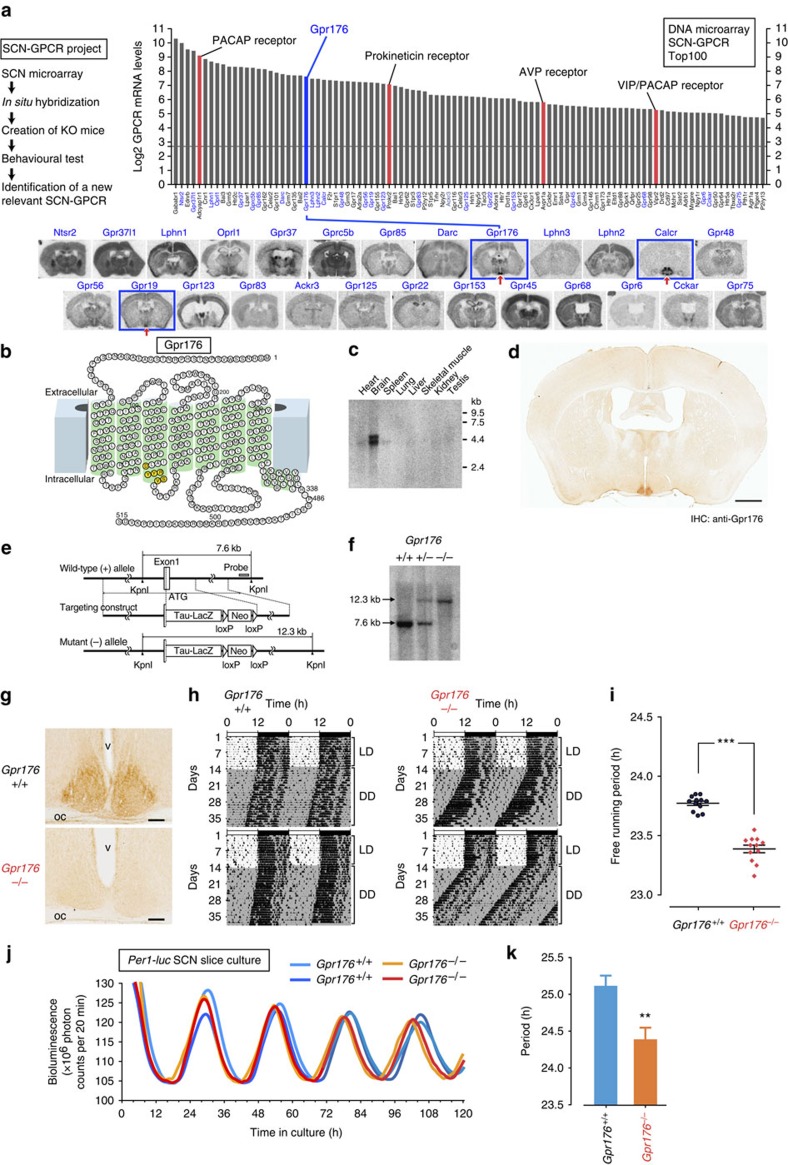
Figure 1. Gpr176 is an SCN-enriched orphan GPCR that sets the pace of circadian timing.
(a) The SCN-GPCR project leading to the identification of Gpr176. The bar graph shows the rank order of expression of the top 100 GPCRs (classes A, B, and C) detected in the SCN microarray analysis (GEO accession number: GSE28574). Red bars are the receptors known to be expressed in the SCN. The horizontal line indicates the threshold of statistical significance of expression. The genes highlighted in blue, which include all listed orphan GPCRs, were characterized further by in situ hybridization using radiolabeled gene-specific probes. Arrows indicate robust positive SCN signals for Gpr176, Calcr and Gpr19. (b) Snake-plot representation of the mouse Gpr176. The residues highlighted in yellow indicate the DRYxxV motif located at the cytoplasmic end of the transmembrane helix III. (c) Northern blotting for Gpr176 with a mouse multiple-tissue blot (Clontech). (d) Representative mouse coronal brain section immunolabeled for Gpr176. Scale bar, 1 mm. (e) Schematic representations of the mouse Gpr176 gene, targeting construct, and the resulting mutant allele. A genomic region downstream of the start codon (ATG) of Gpr176 was deleted. Grey box: probe used for Southern blot. (f) Southern blot of KpnI-digested DNA from Gpr176+/+, Gpr176+/− and Gpr176−/− mice. Genomic fragments from the wild-type (7.8 kb) and mutant (12.3 kb) alleles are indicated. (g) Immunohistochemical confirmation of Gpr176 deficiency in the SCN of Gpr176−/− mice. Scale bar, 100 μm. oc, optic chiasm; v, third ventricle. (h) Representative locomotor activity records of C57BL/6J-backcrossed Gpr176+/+ and Gpr176−/− mice. Mice were housed in LD and then transferred to DD. Periods of darkness are indicated by grey backgrounds. Data are shown in double-plotted format. Each horizontal line represents 48 h; the second 24-h period is plotted to the right and below the first. (i) Circadian periods of free-running activities in DD. Periods of individual mice are plotted. Bars indicate mean±s.e.m. (n=12, for each genotype). ***P<0.001, Student's t-test. (j) Representative Per1-luc bioluminescence records from organotypic SCN slices of Gpr176+/+ (light and dark blue traces) and Gpr176−/− (red and orange traces) mice. (k) Periods of Per1-luc rhythm in Gpr176+/+ and Gpr176−/− SCN slices (means±s.e.m., n=4 for each). **P<0.01, Student's t-test.