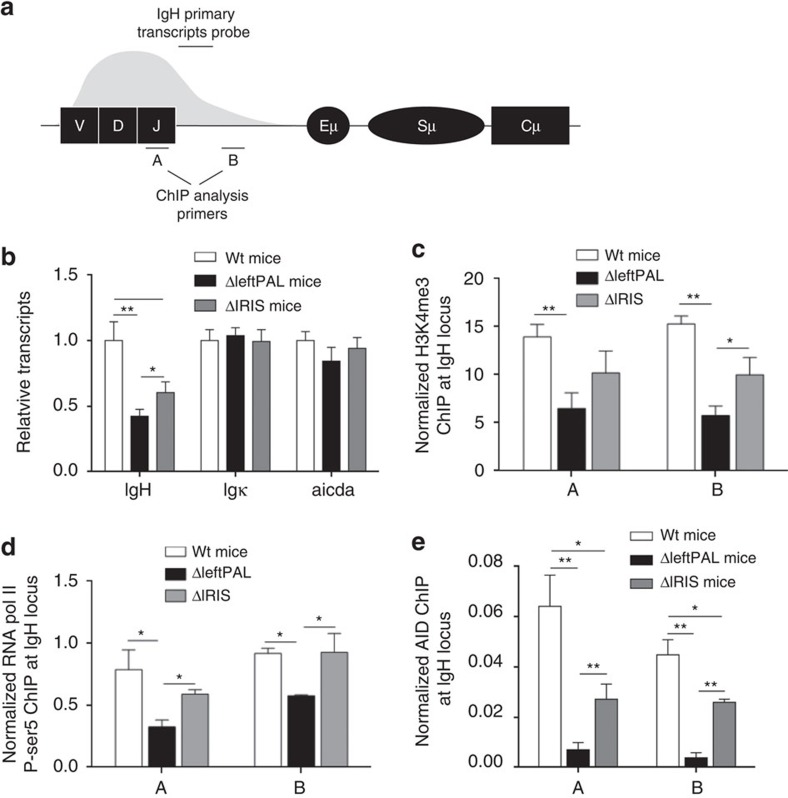
Figure 3. Mechanism underlying the 3'RR palindromic effect on SHM.
(a) Locations of probes (A,B) for ChIP experiments and PCR primers for IgH transcription. (b): IgH, Igκ and AICDA transcription in ΔleftPAL, ΔIRIS and wt mice. Mice were immunised orally with sheep red blood cells for 2 weeks and intraperitoneally with 10 μg of LPS for 3 days. Peyer's patch cell RNA was extracted and transcripts were amplified by real-time PCR. Data are the mean±s.e.m. of six independent experiments with one mouse per genotype (8–12 weeks old, male and female). *P<0.05 and **P<0.001 (Mann–Whitney's U-test for significance). Values were normalised to GAPDH transcripts. (c) ChIP analysis of H3K4me3 in VH regions in Peyer's patch cells in ΔleftPAL, ΔIRIS and wt mice. Background ChIP signals from mock samples with irrelevant antibody were subtracted. ChIP values were normalised to the total input DNA. Data are the mean±s.e.m. of four independent experiments (8- to 12-week-old mice, male and female). *P<0.05, **P<0.01 (Mann–Whitney U-test). ChIP experiments were done in A and B locations (as in a). Same immunisation protocol as in b. (d) ChIP analysis of pol II paused in VH regions in Peyer's patches cells in ΔleftPAL, ΔIRIS and wt mice. Data are the mean±s.e.m. of four independent experiments (8- to 12-week-old mice, male and female). *P<0.05, **P<0.01 (Mann–Whitney U-test). Same immunisation protocol as in b. (e) ChIP analysis of AID recruitment in VH regions in Peyer's patch cells in ΔleftPAL, ΔIRIS and wt mice. Data are the mean±s.e.m. of five independent experiments (8- to 12-week-old mice, male and female). *P<0.05, **P<0.01 (Mann–Whitney U-test). Same immunisation protocol as in b.