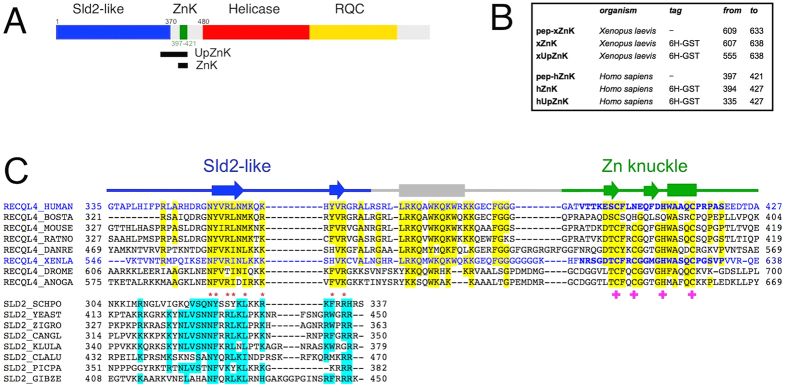
Figure 1. Analysis of RecQ4 N-terminal region.
(A) Structural organization of human RecQ4: the extended Sld2 homology region (blue), the Zn knuckle (green), the helicase core (red) and the RecQ C-terminal domain (RQC, in yellow). The two black rectangles indicate the regions encompassed by the protein constructs made for this study (ZnK and UpZnK). (B) Details of the synthesised peptides (pep-xZnK, pep-hZnK) and recombinant protein fragments (xZnK, hZnK, xUpZnK, hUpZnK) analysed in the present study. (C) Sequence alignment between RecQ4 and the C-terminus of Sld2 proteins. The human and Xenopus sequences are coloured in blue and the residues of the two synthetic peptides pep-hZnK and pep-xZnK are in bold. Residues that are conserved in 6/8 sequences are highlighted in yellow and cyan in RecQ4 and Sld2 proteins, respectively. Residues such as R/K/H, D/E, S/P/T/C, A/G, Y/F/W, I/L/V/M and Q/N, are classified as conserved. Residues conserved across RecQ4 and Sld2 are indicated by red asterisks. Amino acids involved in Zn2+ coordination are indicated by pink crosses. For the Zn knuckle the secondary structure elements (α-helices as rods and β-strands as arrows) are based on the Xenopus NMR structure (PDB ID: 2MPJ), otherwise they are based on the PsiPred prediction (http://bioinf.cs.ucl.ac.uk/psipred/).