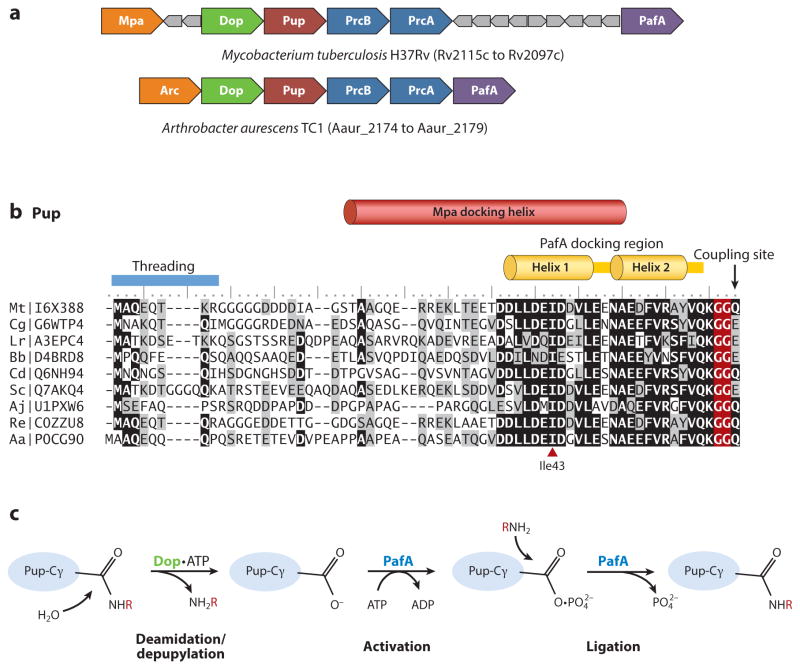
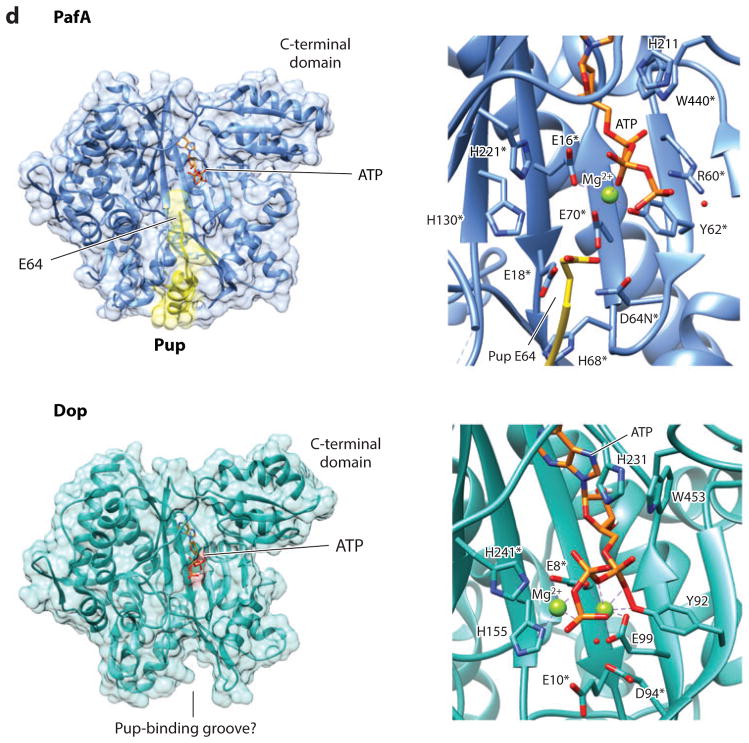
Figure 1.
Pupylation and formation of ubiquitin-like isopeptide bonds. (a) Pup gene neighborhoods. Pup-, Dop-, Mpa-, and 20S proteasome (PrcA/B)-encoding genes are organized in gene neighborhoods in genomes of the phyla Nitrospirae and Actinobacteria. Genes are depicted as arrows, with the arrowheads pointing in the 5′ to 3′ direction. Representative gene neighborhoods are labeled below, including the name of the organism and gene locus tag numbers. (b) Multiple amino acid sequence alignment of representative Pup proteins. Identical and similar residues are highlighted in black and gray, respectively. UniProtKB/TrEMBL accession numbers are included on the left. Critical regions required for Pup activity as described in the text are indicated. (c) Schematic representation of reactions catalyzed by Dop and PafA. Dop mediates the deamidation of Pup-GGQ as well as the depupylation of pupylated proteins, whereas PafA activates and ligates Pup-GGE to target lysines. (d) PafA ligase and Dop depupylase/deamidase are structurally related to carboxylate-amine ligases. PafA and Dop structures are based on Protein Data Bank ID 4BJR and 4B0S, respectively. The Pup C-terminal tail that lines a groove on the surface of PafA that leads to the putative active site is highlighted in yellow. Mg-ATP and residues (*) required for catalytic activity based on site-directed mutagenesis and substrate analog trap are also indicated.