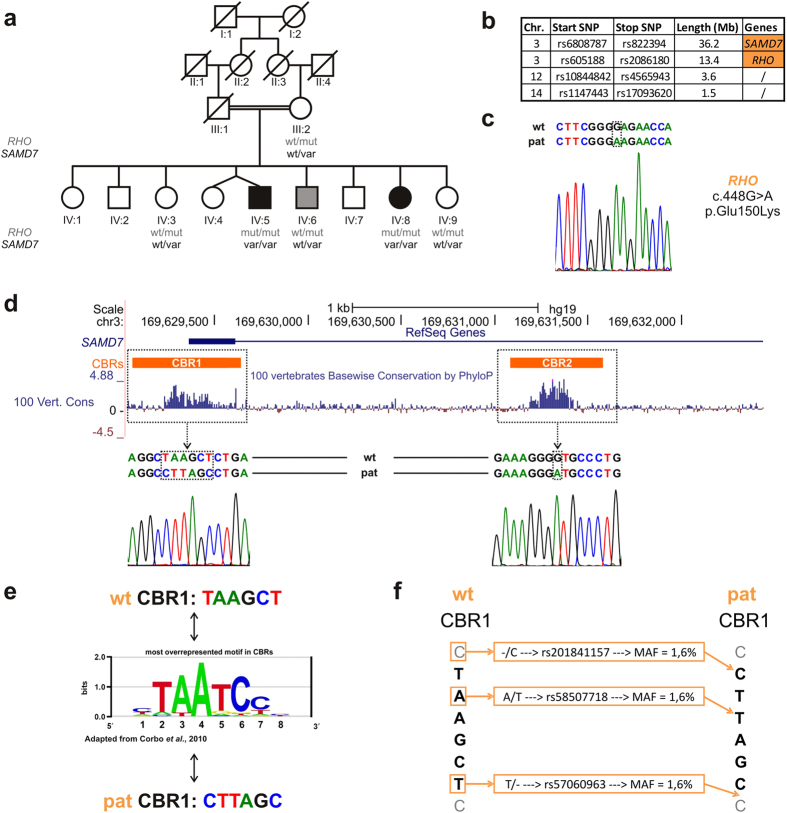
Figure 1. Identification of a homozygous RHO mutation and non-coding SAMD7 variants.
(a) Pedigree of the family. Six family members participated in this study and were clinically and genetically investigated. The genotype of each individual for the RHO mutation is indicated in grey, while the genotype for the SAMD7 variants is shown in black. The affected siblings IV:5 and IV:8 are homozygous for both variants, while all other family members are heterozygous carriers. One of these siblings IV:6 displays minor subclinical manifestations, more specifically pigmentary anomalies on fundus photography and subjective complaints of night blindness. (b) Homozygosity mapping. SNP chip analysis in the affected siblings revealed four shared homozygous regions, harboring the SAMD7 and the RHO genes. Both genes are located on chr. 3, but are not genetically linked as they are 40 Mb apart. (c) RHO mutation. Electropherogram showing the homozygous RHO c.448G > A, p.(Glu150Lys) identified in the affected siblings. Segregation of this mutation is depicted in Fig. 1a. (d) SAMD7-associated regulatory variants. UCSC genome browser view of the SAMD7 locus (http://genome.ucsc.edu/). The horizontal blue line in the upper part of the figure represents the structure of the SAMD7 gene, here only showing the first non-coding exon and a part of the adjacent intron. The orange blocks indicate the SAMD7-associated CBRs, with CBR1 located in the promoter region and CBR2 in the first intron. The next track represents the PhyloP conservation scores across 100 vertebrates, showing high conservation of both CBRs. The electropherograms show the homozygous variants located in the CBRs identified in both affected siblings. Segregation analysis of these variants is represented in Fig. 1a. (e) Disruption of CRX-binding motif. The middle part depicts the most overrepresented motif in CBRs, as reported by Corbo et al.17 While the wt CBR1 sequence is very similar to this CRX-binding motif, the variants identified in this study seem to disrupt it completely. (f) CBR1 variation. Comparison of the wt and the variant sequences shows that the variation in CBR1 consists of three SNPs with MAF 1.6%: rs201841157, rs58507718 and rs57060963 leading to an insertion, transversion and deletion event, respectively.