Fig. 7.
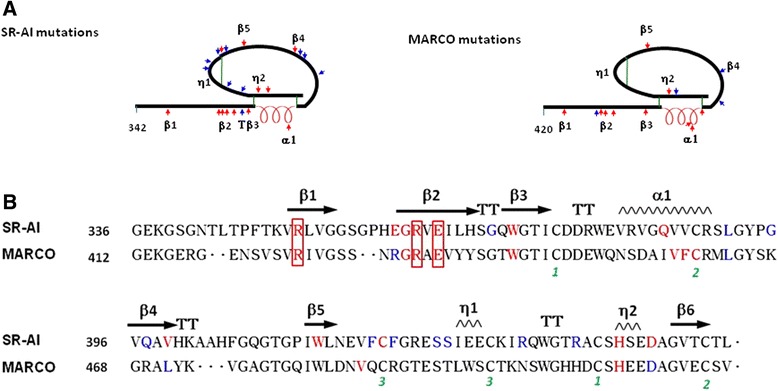
Comparison of the amino acid required for targeting surface and internalizing Aβ between SR-AI and MARCO. a Schematic representation of SRCR domain of SR-AI and MARCO and the mutation sites are shown. The surface targeted mutations are shown as blue arrows and the intracellular retained mutantions are shown as red arrows. The black line indicates the sequence of SRCR; α-helix is indicated as red spring, and disulfide bonds are shown as green line. The secondary structure of the SRCR domain of SR-AI, β-strands (β1-6), α-helix (α) and 310 helices (η1, η2) are indicated. b Sequence alignment of human MACRO and SR-AI using ClustalW2 and ESPrip. The β-sheets are shown as arrows, and helixes are shown as a saw tooth pattern. Turns are marked as T. The three pairs of disulfide bonds are labeled as numbers in green. The amino acid in red indicates its mutation is not surface targetable. The amino acid in blue indicates its mutation is surface targetable and be able to internalize oAβ
